Research Article - Clinical Practice (2017) Volume 14, Issue 2
Effect of four intestinal strains on TNF-a ̱̉ IL8 and IL6 expression in Caco-2 cells
- Corresponding Author:
- Yongbo Kang
Kunming University of Science and Technology
Kunming, Yunnan, China
E-mail: 657151276@qq.com
Abstract
Gut microbiota play an important roles in induction of various cellular responses, including cytokine production. In this work, we examined the involvement of four intestinal strains in induction of the cytokine response (the relative expression of TNF-α, IL-6, and IL-8) in intestinal epithelial cells. Caco-2 cells were treated with cell-free supernatants (CFS) of four intestinal strains: Shigella spp., Bacteroides fragilis, Weissella cibaria, Enterococcus faecium. Shigella spp., Bacteroides fragilis, Weissella cibaria, Enterococcus faecium CFSs resulted in marked an increase of mRNA TNF-α content ( p<0.05 or p<0.01).In additon, Shigella spp. CFS, Bacteroides fragilis CFS, Weissella cibaria CFS significantly increased expression mRNA for IL-8 ( p<0.05 or p<0.01), In contrast, Enterococcus faecium caused significant a decrease of mRNA IL-8(p<0.01). Last but not the least important, Shigella spp., Bacteroides fragilis, Weissella cibaria, Enterococcus faecium CFSs led to mark an increase of mRNA IL-6 level (p<0.01). These results indicate that four intestinal strains produce bioactive metabolites, suggesting that gut microbiota metabolites may therefore provide an entirely new approach towards protection of intestinal epithelial and treatment of human diseases.
Keywords
gut microbiota, Caco-2, metabolites, cytokine response, TNF-α, IL-6, IL-8
Introduction
The microflora of adult humans is found primarily in the colon and distal small intestine and consists of α1013 microorganisms, comprising nearly 500 species and nearly 2 million genes (the “microbiome”) [1]. The intestinal mucosal surface is a primary component of the host defense against potentially harmful microorganisms. Simultaneously, this surface is also the location where the intestinal microbiota and the host coexist in a tightly regulated balance. Evidence is emerging for interactions, termed “cross talk,” between the intestinal microflora and the diverse population of cells in the intestinal mucosa that result in several physiological and immunologic responses [2-5].The intestinal epithelium synthesizes many mediators that are involved in the innate immune response. Whether the response is pro- or antiinflammatory may depend on the balance of individual microorganisms that colonize the intestinal lumen at any particular time. Host identification of microbial components is vital for the development of innate and adaptive immune responses to gastrointestinal infections. Furthermore, both innate receptors and microbes have been identified as important players in the balance between the induction and prevention of autoimmunity in the host [6-9]. Controversy exists as to whether contact between an intestinal bacterial cell and an epithelial cell in the gut is needed to confer beneficial or harmful effects (the cytokine response) [10], or whether metabolites from gut bacteria are sufficient to cause this effect. To address this question, in this study, we tested in Caco-2 intestinal epithelial cells whether metabolites from four intestinal strain (Shigella spp., Bacteroides fragilis, Weissella cibaria, Enterococcus faecium) are sufficient to affect TNF-α interleukin-8(IL8) and interleukin-6(IL6) expression.
Materials and Methodology
Isolation of gut bacteria
Faecal samples were provided by 2 healthy adults (aged 20-40) who had not been prescribed antibiotics for at least 3 months prior to this study nor had taken any probiotics. For the isolation of gut bacteria, freshly collected faecal samples were suspended in phosphate-buffered saline, and after homogenization, serial dilutions were performed from these suspensions by plating to de Man–Rogosa–Sharpe (MRS) agars and incubation of the plates were conducted at 37°C under anaerobic conditions for 48 h. From these plates, potential different isolates were selected and further subcultured in MRS broth and then stored at –80°C in glycerol (40% v/v).
Bacterial identification by 16S rRNA gene sequencing
All isolates were genotypically discriminated by rep-PCR as described previously [11], and selected strains were identified. The ~1.5 kb 16S rRNA gene of each of the selected strains was amplified with primers AMP_F(5′-GAGAGTTTGATYCTGG CTCAG-3′) and AMP_R(5′- AAGGAGGTGATCCARCCGCA-3′)[12]. PCR mixtures contained 1 μL of DNA template from genomic DNA extracted from each strain using a DNA Isolation kit (Qiagen, Germany), 45 μL of 1.1 × PCR Mix(Promega, China), 1 μL of 10 μM primers AMP_F and AMP_R. PCR was performed using a thermal cycler (Benchmark, TC9639) with the following programme: 98°C for 2 min, 30 cycles of 98°C for 10 s, 55°C for 15 s, and 72°C for 30 s, and 72°C for 5 min final extension. PCR products were run on a gel to check the amplification, and amplicons were further purified using a Sure-Clean kit (Bioline). Sequencing reactions were prepared using primers AMP_F and AMP_R at 1.6 μmol/L concentrations and the ABI Prism BigDye Terminator version 3.1 Cycle Sequence kit (Applied Biosystems) according to the manufacturer’s protocol. Sequences obtained were interrogated in the NCBI database by the BLAST algorithm with a similarity criterion of 97%-100%.
Caco-2 cell culture
Caco2 cell line (Human Caucasian colon adenocarcinoma; ECACC) was cultured in Dulbecco’s modified Eagle’s medium (DMEM; Lonza, Verviers, Belgium) supplemented with with 10% FBS (Integro BV, Zaandam, The Netherlands) and 100 μ/ml of Penicillin– Streptomycin (Gibco, Grand Island,USA) solution. Cells were maintained in 5% CO2 at 37° in a humidified incubator.
Cultivation of bacteria
Isolated Shigella spp., Bacteroides fragilis, Weissella cibaria, Enterococcus faecium were cultivated an anaerobically at 37° in Man, Rogosa and Sharpe (MRS) broth until OD600=0.6-0.7. Cell-free supernatants (CFSs) were collected by centrifugation (25°, 5 min, 3000 × g) and stored at 20° until used in the experiments.
Experiments with Caco-2 cells
Caco-2 cells in T25 flask were washed and suspended, and subsequently into a 6-well plate (4 × 105 cells/well). When cell confluence was approximately 80%, culture medium was discarded and cell monolayers were washed twice with PBS. CFS was sterilized through 0.2 mm sterile syringe filter units (Sartorius, Goettingen, Germany) were performed. Sterile CFS was diluted 10% (v/v) in DMEM medium. Then, cells were added with 2ml DMEM medium containing10% (v/v) sterile CFS incubated at 37° with 5% CO2 for 24 h.
RNA isolation and real-time quantitative PCR
Total RNA was isolated using a Machery- NucleoSpin RNA II kit (Bioke, Leiden, Netherlands) and quantified using a Nanodrop ND-1000 (Wilmington, DE, USA). cDNA was prepared from total RNA using a cDNA Synthesis kit (TAKARA BIO INC.) with random primers. Real-time PCR reactions (50° for 2 min, 95° for 10 min, followed by 50 or 60 cycles of 95° for 15 s and 58° for 30 s and 72° for 10 min) were performed with SYBR Green-based real-time PCR (Applied Biosystems, Austin, USA) according to the manufacturer’s instruction. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene was used as reference. Relative gene expressions were normalized to GAPDH using the formula 2- ∇ ∇ CT (- ∇ ∇CT= ∇CT sample - ∇CT control). The sequences of the oligonucleotide primers used were as follows: 5′-GTACATCCTCGACGGCAT CTC-3′ and 5′-TGTGGTTGGGTCAGGG GTGGT-3′ for IL-6, 5′-CTGGCCGTGGCTCTCTTGGCAGCCTTCCTG- 3′ and 5′-GGCAACCCTACAACAGACCCACACAATACA- 3′ for IL-8, 5′-TCTCGAACC CCGAGTGACAA-3′ and 5′-TATCTCTCAGCTCCACGCCA- 3′ for TNF-′ and 5′-C GAGATCCCTCCAAAATCAAG-3′ and 5′-GAGCTTGACAAAGTGGTCG-3′ for GAPDH.
Statistical analyses
The statistical significance of differences between treatments was determined using ANOVA and the two-tailed Student’s t-test and p-values of 0.05 or less were considered significant. All comparisons were made against the respective differentiation medium (DM)-only controls unless otherwise stated.
Results
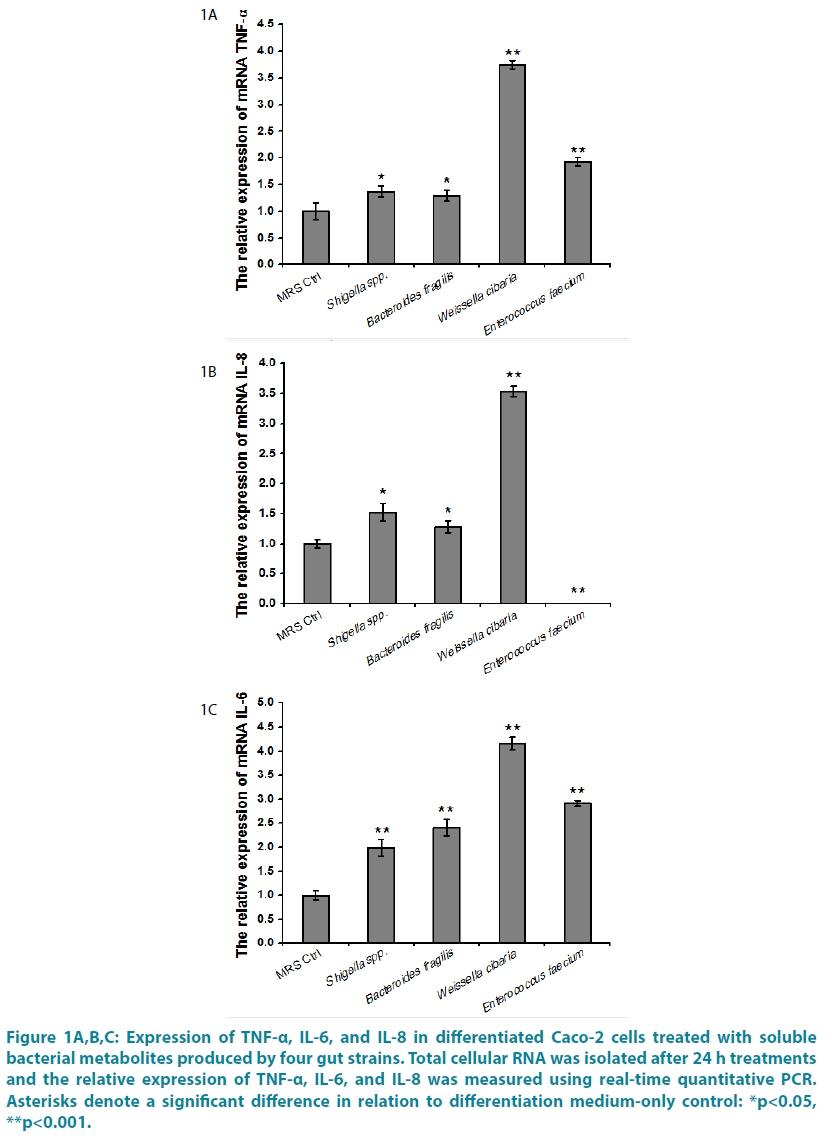
The relative expression of TNF-α, IL-6, and IL-8 in Caco-2 cells treated with four gut bacteria CFSs.
We investigated the effect of four gut bacteria CFSs secreted factors by treating Caco-2 cells for 24 h with Shigella spp., Bacteroides fragilis, Weissella cibaria, Enterococcus faecium CFSs. The relative expression of TNF-α, IL-6, and IL-8 were studied. Shigella spp., Bacteroides fragilis, Weissella cibaria, Enterococcus faecium CFSs resulted in marked an increase of mRNA TNF-α content ( p<0.05 or p<0.01) (FIGURE 1A). TNF-α plays a central role in the intestinal inflammation of various inflammatory disorders including Crohn’s disease (CD). TNF-α-induced increase in intestinal epithelial tight junction (TJ) permeability has been proposed as one of the proinflammatory mechanisms contributing to the intestinal inflammation [13-15]. Furthermore, the proposed proinflammatory actions of TNF-α include production and stimulation of proinflammatory cytokines including IL-1, -6, -8, -12 [15]. In additon, Shigella spp. CFS, Bacteroides fragilis CFS, Weissella cibaria CFS significantly increased expression mRNA for IL-8 (p<0.05 or p<0.01). In contrast, Enterococcus faecium caused significant a decrease of mRNA IL-8 (p<0.01) (FIGURE 1B). IL-8 is produced through cell receptors that interact with the nuclear factor- κB (NF-κB) pathway under the action of several proinflammatory mediators such as Escherichia coli lipopolysaccharide (LPS), flagellin, and TNF-α. High concentrations of IL-8 are thought to be involved in the pathogenesis of several diseases including neonatal necrotizing enterocolitis [16,17]. Last but not the least important, Shigella spp., Bacteroides fragilis, Weissella cibaria, Enterococcus faecium CFSs led to marked an increase of mRNA IL-6 level (p<0.01) (FIGURE 1C). In the case of Caco- 2 cells, it has been reported that IL-6 induced the activation of NF-_B in Caco2-BBE cells and resulted in the expression of intracellular adhesion molecule 1, which plays an important role in inflammation through some form of leukocyte-epithelial cell adhesion [18]. In a previous study, environmental conditions, such as pH, were shown to be able to affect NFκB, which regulates the transcription of a large number of genes involved in inflammation and immune responses, including those coding for gamma interferon (IFN-β), IL-1β, IL-2, IL-6, IL-8, TNF-α, and granulocyte /macrophage or granulocyte/ colony-stimulating factor [19].
Figure 1: Expression of TNF-α, IL-6, and IL-8 in differentiated Caco-2 cells treated with soluble bacterial metabolites produced by four gut strains. Total cellular RNA was isolated after 24 h treatments and the relative expression of TNF-α, IL-6, and IL-8 was measured using real-time quantitative PCR. Asterisks denote a significant difference in relation to differentiation medium-only control: *p<0.05, **p<0.001.
Discussion and Conclusion
Both live and heat-killed gut microbiota have been shown to the cytokine response in Caco-2 cells [20]. Similar results can be obtained with CFSs devoid of bacterial cells. Regulation of the cytokine response are observable depending on the bacterial CFS used, implying that there are differences in the components secreted between different species and strains. Similarly, the team of Putaala et al. [21] also found that bacterial CFSs can affect tight junction integrity on Caco- 2 cells and inflammatory reactions (expression of COX 1, 2, and 3). In conclusion, direct contact between epithelial cells and gut bacteria may not always be necessary for biological effects, as cytokine response could be mediated by secreted components of specific strains. It remains to be investigated whether the active metabolites are produced in fermented probiotic products or in situ in the intestine, and whether they mediate similar effects in vivo. Intestinal microbiota metabolites may therefore provide an entirely new approach towards treatment of human diseases. Research has just started in this field, and more research is urgently required.
Ethics Approval
Fecal samples were taken after informed written consent, in accordance with guidelines endorsed by the Chinese human ethics committee.
Conflict of Interest
The authors declare no conflicts of interest.
Acknowledgements
We are grateful to Yue Cai for help in cultivation of the microbes, cell culture experiments and gene expression analysis
References
- Xu J, Gordon JI. Honor thy symbionts. Proceedings of the National Academy of Sciences of the United States of America 100(18):10452 (2003).
- Bourlioux P, Koletzko B, Guarner F, et al. The intestine and its microflora are partners for the protection of the host: report on the Danone Symposium “The Intelligent Intestine,” Paris 2002. JSci Food Agric69(4):495–500 (1995).
- Hooper LV, Midtvedt T, Gordon JI. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr22(1):283-307 (2002).
- Haller D. Non-pathogenic bacteria elicit a differential cytokine response by intestinal epithelial cell/leucocyte co-cultures. Gut 47(1):79-87 (2000).
- Walker WA. Role of nutrients and bacterial colonization in the development of intestinal host defense. JPediatrGastroenterolNutr30 Suppl 2 (Suppl 2):S2 (2000).
- Kang Y, Zhang X, Cai Y, et al. Gut microbiota and metabolic disease: from pathogenesis to new therapeutic strategies. RevMed Microbiol 27(4):141-152(2016).
- Kang YB, Cai Y, Zhang H. Gut microbiota and allergy/asthma: From pathogenesis to new therapeutic strategies. AllergolImmunopathol (Madr)2016.
- Kang Y, Cai Y, Zhang X, et al. Altered gut microbiota in RA: implications for treatment. Z Rheumatol1-7 (2016).
- Kang Y, Kong X, Zhang X, et al. Progress in research on interaction of intestinal microbiota and immunity. J Zhejiang Univ2016.
- Karczewski J, Groot J. Molecular physiology and pathophysiology of tight junctions III. Tight junction regulation by intracellular messengers: differences in response within and between epithelia. Am J PhysiolGastrointest Liver Physiol279(4):G660 (2000).
- Sagdic O, Ozturk I, Yapar N, et al. Diversity and probiotic potentials of lactic acid bacteria isolated from gilaburu, a traditional Turkish fermented European cranberrybush( Viburnumopulus L.) fruit drink. Food Res International64:537-545 (2014).
- Baker GC, Smith JJ, Cowan DA. Review and re-analysis of domain-specific 16S primers. J Microbiol Methods 55(3):541-555 (2003).
- Murch SH, Braegger CP, Walker-Smith JA, et al. Murch, SH, Braegger, CP, Walker-Smith, JA and MacDonald, TT. Location of tumour necrosis factor α by immunohistochemistry in chronic inflammatory bowel disease. Gut 34: 1705-1709. Gut 34(12):1705-1709 (1994).
- Ricart E, Panaccione R, Loftus EV, et al. Infliximab for Crohn's disease in clinical practice at the Mayo Clinic: the first 100 patients. Am J Gastroenterol 96(3):722-729 (2001).
- Sands BE. Biologic therapy for inflammatory bowel disease. Inflammatory Bowel Diseases 3(2):95 (1997).
- Nanthakumar NN, Fusunyan RD, Sanderson I, et al. Inflammation in the developing human intestine: A possible pathophysiologic contribution to necrotizing enterocolitis. Proceedings of the National Academy of Sciences of the United States of America 97(11):6043-6048 (2000).
- Edelson MB, Bagwell CE, Rozycki HJ. Circulating pro- and counterinflammatory cytokine levels and severity in necrotizing enterocolitis. Pediatr 103(1):766-771 (1999).
- Wang L, Walia B, Evans J, et al. IL-6 induces NF-kappa B activation in the intestinal epithelia. J Immunol 171(6):3194-3201 (2003).
- Kopp EB, Ghosh S. NF-kappa B and rel proteins in innate immunity. AdvImmunol58(1):1-27 (1995).
- Klingberg TD, Pedersen MH, Cencic A, et al. Application of Measurements of Transepithelial Electrical Resistance of Intestinal Epithelial Cell Monolayers To Evaluate Probiotic Activity. ApplEnviron Microbiol 71(11):7528 (2005).
- Putaala H, Salusjärvi T, Nordström M, et al. Effect of four probiotic strains and Escherichia coli O157:H7 on tight junction integrity and cyclo-oxygenase expression. ResMicrobiol 159(9-10):692 (2008).