Research Article - Clinical Investigation (2022) Volume 12, Issue 2
Encapsulation of Remdesivir with NV-COV-2 biopolymer delays formation of GS-441524 in vivo
- Corresponding Author:
- Ashok Chakraborty
Allexcel, Inc., 1, Controls Drive, Shelton, CT, USA
E-mail: ashok.chakraborty@allexcel.com
Received: 10-February-2022, Manuscript No. fmci-22-54054; Editor assigned: 12- February-2022, PreQC No. fmci-22-54054 (PQ); Reviewed: 21-Febuary-2022, QC No. fmci-22-54054 (Q); Revised: 23-Februay-2022, Manuscript No. fmci-22-54054 (R); Published: 28-February-2022, DOI: 10.37532/2041-6792.2022.12(2).23-29
Abstract
Among all the promising antiviral drugs, so far approved by FDA for using against SARS-CoV2 RNA viruses is Remdesivir (Gilead sciences). It is an adenosine nucleoside monophosphate, actually a prodrug GS-5734 previously been used against Ebola in rhesus monkeys. Here we will present the pharmaco kinetics of GS 441524 in vivo; and also the effect of encapsulation with our biopolymer NV-CoV-2 in there.
Keywords
Remdesivir • GS-441524 • SARS-CoV-2 • Pharmacokinetics • NV-CoV-2 • Nanoviricide • Biopolymer • Rat • In vivo
Introduction
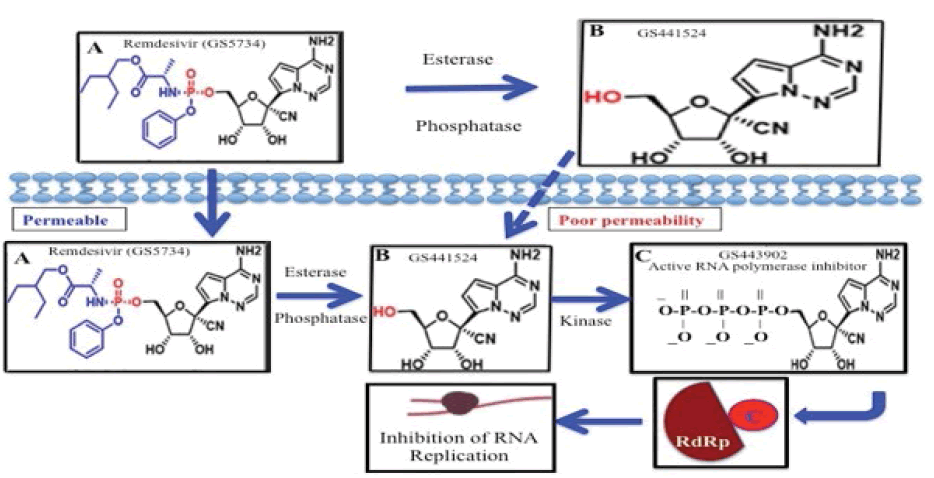
Remdesivir, GS-5734, is a prodrug containing monophospho unit in its adenosine analogue, GS-441524 which is converted intracellularly to an active nucleoside triphosphate derivative (GS-443902) that inhibits viral RNA polymerases (RdRp) but does not affect host RNA or DNA polymerases (Figure 1) [1-6].
Remdesivir (RDV) has a broad action against RNA viruses, including Ebola, SARS, MERS [7-13]. Very recent, WHO has declared Remedisvir to be used against COVID-19 disease [12,13]. Evaluation of intravenously administered Remdesivir tolerance and safety in 94 healthy adult volunteers has found it to be generally well-tolerated and to have an acceptable safety profile [14].
GS-441524 is the Predominant Metabolite in the Bloodstream when Remdesivir is Administered IV
Hydrolytic enzymes, esterases and phosphatases are ubiquitous in serum, and those bio-activates the prodrug to its active derivatives in vivo [15,16]. For example, PK of Remdesivir in Nonhuman Primates (NHP) have concluded that intact Remdesivir exhibits a short plasma half-life of about 0.4 h in serum with the appearance of nucleoside, GS-441524 after IV injection of the prodrug in NHP [4,17]. However, the wide range of EC50 values of Remdesivir in vitro can be explained by the differential expression of bioactivating enzymes, such as esterase, phosphatase [7,18,19].
Remdesivir vs. GS-441524 for antiviral application
While Remdesivir has shown some efficacy in patients with advanced COVID-19, its phosphate prodrug GS-441524 shows comparable efficacy in cell-based models of primary human lung and cat cells infected with coronavirus [12,20]. In humans, Remdesivir showed moderate protein binding (80%-90%) and GS-441524 exhibited very low protein binding in plasma (<20%) [21]. The half-lives are about 0.89 h and 25 h for Remdesivir and GS-441524, respectively. Remdesivir is mainly excreted in urine (about 74%) in the form of GS-441524 which can challenge the efficacy of the drug against the disease treatment.
Our study aimed to investigate the pharmacokinetics of GS- 441524 in plasma after intravenous administration of RDV alone and when biopolymer-protected in a virus-infected rat model.
Methodology
Materials and Methods: Test Articles
• NV1067-387: Polymer
• NV1067-387-R: Polymer encapsulated RDV
• NV1067-376: RDV in sulfobutylether-β-cyclodextrin sodium (SBECD, Gilead)
• RDV in DMSO: MeOH
In vivo treatment
Eighteen Sprague Dawley female rats (Taconic Biosciences, USA) (three/each control and in treatment groups) were administered with NV-CoV-2 or NV-CoV-2R, once per day for 5 days (0, 1, 3, 5, and 7) over 7 days. DMSO was used as a vehicle for the control group. Injection on day 0 is considered as a 1st injection and on day 7 is the 5th injection. NV-376 (RDV-in-SBECD; Gilead) was given in two doses on day 1 followed by a daily dose through day 7. Each compound was delivered via slow-push IV injection. Blood samples for systemic exposure assay were collected at 0, 0.08, 0.5, 1, 2, 4, 8 and 24 hours after 1st and 5th injection of the drugs.
Blood samples were taken from one animal per sex in each test article treated group at each time point. The same animal was used to draw blood across all the time points after “day 0” and “day 7” injections.
The procedures for in vivo experiments were done by Dr Krishna Menon from AR Biosystems, (17633 Gunn Highway, Odessa, FL 33556), based on the protocol #IACUC No. 14/17ARB. The study design is shown below in Table 1.
| Group | Drug | Dose (mg/Kg) | Dose Volume (mL/Kg) | Concentration (mg/mL) NV-CoV-2/Remdesivir |
|---|---|---|---|---|
| 1 | NV-1067-387-High | 320 | 10 | 32/0 |
| 2 | NV-1067-387-Medium | 160 | 10 | 16/0 |
| 3 | NV-1067-387-R-Low | 80 of NV-CoV-2, plus 8 of Remdesivir | 10 | 8/0.8 |
| 4 | NV-1067-387-R-Medium | 160 of NV-CoV-2, plus 16 of Remdesivir | 10 | 16/1.6 |
| 5 | NV-1067-376 | 0 of NV-CoV-2, plus 16 of Remdesivir in SBECD (Gilead) | 10 | 0/1.0 |
| 6 | NV-1067-377 | Vehicle (1xPBS) | 10 | 0/0 |
Table 1. Study design
Preparation of standard curve of Remdesivir and GS-144524
Reagents are procured from different vendors as shown in Table 2. Standard Curves Preparation of Remdesivir and GS-441524 were determined by using LC-MS at different concentrations of the standard solutions in DMSO+MeOH (1:9), ranging from 0 ng/uL-5 ng/uL. Plasma and/or PBS was used in the reaction mixture. As an Internal Standard (ISTD), 13C6-RDV and 13C5-GS441524 (2.5 ng/uL) were used, respectively for RDV and GS assay. The final concentration of the internal standards in the reaction mixture becomes 0.125 ng/uL. Extraction of RDV for LC-MS assay was done by using an Acetonitrile cocktail solution (Acetonitrile: ISTDs: 75% Isopropanol at 10:1:1), added at the ratio of 1:4 by volume. The mixture was mixed well and centrifuged for about 5 minutes at 10,000 RPM to separate the precipitated solids from the supernatants. Concentrations of RDV and GS-441524 compared to their ratio with the ISTDs (13C6-RDV, 13C5-GS, respectively) were used to generate a standard curve for RDV and GS- 441524, which were shown in the result section.
| Item(s) | Vendor | Cat# | Lot# | Description |
|---|---|---|---|---|
| PBS 1X, pH 7.4 | Life Technologies | pH 7.2 | ||
| Low-binding tubes | Eppendorf | 20431081 | ||
| Na-Acetate Buffer 1M, pH 5.2 | Sigma | S7899 | SLBS5549 | |
| Acetonitrile for precipitation | 100% | |||
| Rat- Plasma-Sprague Dawley | Innovative Research | IGRTNaEDTA 22443 | Anticoagulant-NaEDTA | |
| Remdesivir (RDV) | Medkoo Biosciences, Inc. | 329511 | E20004S01 | |
| GS-445124 | Medkoo Biosciences | 555299 | C20R07B09 | |
| ISTD (13C6-isotope) for RDV | AlsaCHIM | 8845 | MJ-ALS-20037-P1 | |
| ISTD (13C5-isotope) for GS-441524 | AlsaCHIM | 8855 | MJ-ALS-20050-P1 | |
| Isopropanol | Sigma-Adrich | 75% |
Table 2. Sources of reagents
Assay of Remdesivir and GS-144524 in Rat Plasma after the drug’s injection
To test the in vivo stability of our in-house made anti- COVID-19 product, NV-CoV-2 polymer encapsulated RDV, we have injected our sample along with the polymer only as a negative control, at two different doses (Low, and Medium, see Table 1). As a positive control, we included Gilead RDV compounds in SBECD. After the 1st and 5th injections (day 1 and day 7) of the test materials, blood samples were collected at different time points for 24 hrs. Received samples from the test site were pre-diluted (1:2) and diluted further to (1:20) with plasma:3M Na-acetate, pH 5.0 (1:1). In those samples, reaction mix including ISTDs, 75% Isopropanol and Acetonitrile (1: 1:10) was added at the ratio of 1:4 by volume for the extraction of RDV from the test materials. Vortex and spin the solution for about 5 minutes at 10,000 RPM to separate the precipitated solids from the supernatants. To decrease the plasma esterase activity all steps were performed on ice.
Detection of RDV and GS-441524 by LC-MS Spectroscopy
Analysis was performed using the LC and MS analysis conditions shown in Table 3, and the Multiple Reaction Monitoring (MRM) data acquisition parameters shown in Table 4. Shim-Pack SceptorTM C18120 (50 mm × 2.1 mm I.D., 1.9 uM) was used as the analytical column.
| LC Analysis Conditions | |
|---|---|
| HPLC Method | RDVmetIS_LC_NoSlit-0.4mL-ExtRinse_202010231219.1 cm |
| Column | Shimadzu, Shim Pack, Sceptor, C18-120, 2.1 mm × 50 mm, 1.9 um, Cat#227-31120-01 |
| Pre-Column | Guard Column, Shimadzu, Shim-pack, C18-120 EXP (G), 1.9 um, 2.1 mm × 5 mm, or Restek prefilter: Ultrafield UHPLC, 0.2 um frit, Catalog# 25810. |
| Mobile Phase | A: Water containing 0.05% formic acid |
| B: Acetonitrile containing 0.05% formic acid | |
| Gradient | 0 min-0.3 min: 5% B, |
| 0.3 min-0.35 min: 30% B, | |
| 0.35 min-1.5 min: 70% B, | |
| 1.5 min-1.8 min: 90% B, | |
| 1.8 min-2.8 min: 90% B, | |
| 2.8 min-2.9 min: 5% B, | |
| 2.9 min-4.5 min: 5% B, | |
| (A: Solvent A; B: Solvent B) | |
| Injection Volume | 1 uL |
| Auto-sampler was used in external rinsing mode, with R0=95/5 Water/Acetonitrile and R3=1/1 IPA/Methanol. | |
Table 3. Chromatographic conditions
| Compounds | Ion | Precursor Ion (m/z) | Product Ion (m/z) |
|---|---|---|---|
| RDV (target, drug) | Quantitation ion | 603.2 | 200.0 (@41) |
| Quantitation ion | 603.2 | 229.0 (@21) | |
| 13C6-RDV (Internal standard for RDV) | Quantitation ion | 609.2 | 206.0 (@25) |
| Quantitation ion | 609.2 | 229.0 (@26) | |
| GS-441524 (target, metabolite) | Quantitation ion | 292.1 | 163.0 (@25) |
| Quantitation ion | 292.1 | 136.0 (@26) | |
| 13C5-GS-441524 (Internal standard for metabolite) | Quantitation ion | 297.1 | 204.1 (@25) |
| Quantitation ion | 297.1 | 148.0 (@30) |
Table 4. MS MRM Transitions observed (for quantitation)
Calculation of RDV and GS-441524 from the Chromatogram
From the chromatogram, the ratio of RDV and its isotope, 13C6-ISTD (as an internal standard) was calculated. The amount of RDV was determined using the linear equation derived from their respective standard curve.
The ratio of RDV and its isotope, 13C6-ISTD (as an internal standard) was calculated. Similarly, the ratio of GS and its internal standard, 13C5-ISTD was also calculated. The amount of RDV and GS were determined using the linear equation derived from their respective standard curve. The values were normalized using the dilution factor (2 × 20 × 4=160) used for the original plasma sample and expressed as the ratio of RDV/GS-441524 found in plasma (ug/mL) and the amount of RDV administered (mg/kg of rat body weight), as described elsewhere [22].
Ethical Statement
All the animal handling experiments are done by Dr Krishna Menon from AR Biosystems, (17633 Gunn Highway, Odessa, FL 33556), as per the protocol #IACUC No. 14/17ARB. In brief, there are no known acceptable alternatives to the use of live animals to accomplish the purpose of this study. The animal is a standard model for non-clinical studies, for which there is a large historical database, and has been found a suitable model in previous investigations with the same disease models.
This study was designed to use the fewest number of animals possible consistent with the scientific objectives of the study, the contemporary scientific standards, and in consideration of applicable regulatory requirements. The number of animals used is considered the minimum to make meaningful biological comparisons and for statistical calculations.
Results
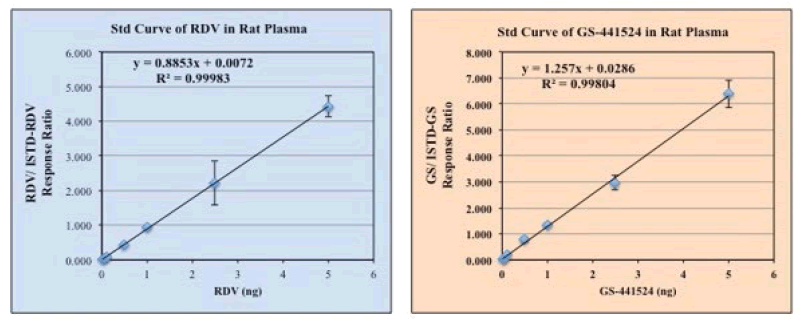
A standard curve for RDV and GS-441524 from a representative experiment were shown in Figure 2.
Figure 2: Standard Curve of Remdesivir and GS-441524 were determined by using LC-MS, using different concentrations of the standards solution in DMSO+MeOH (1:9) ranging from 0 ug/mL-5 ug/mL final concentrations. Plasma and/or PBS were used in the reaction mixture when needed. As an internal standard (ISTD) 13C6-RDV and 13C5-GS, 2.5 ug/mL each, were used, respectively for RDV and GS assay, and or in the mixture. Final extraction of RDV and its metabolite GS was done by using acetonitrile
Pharmacokinetics of Remdesivir
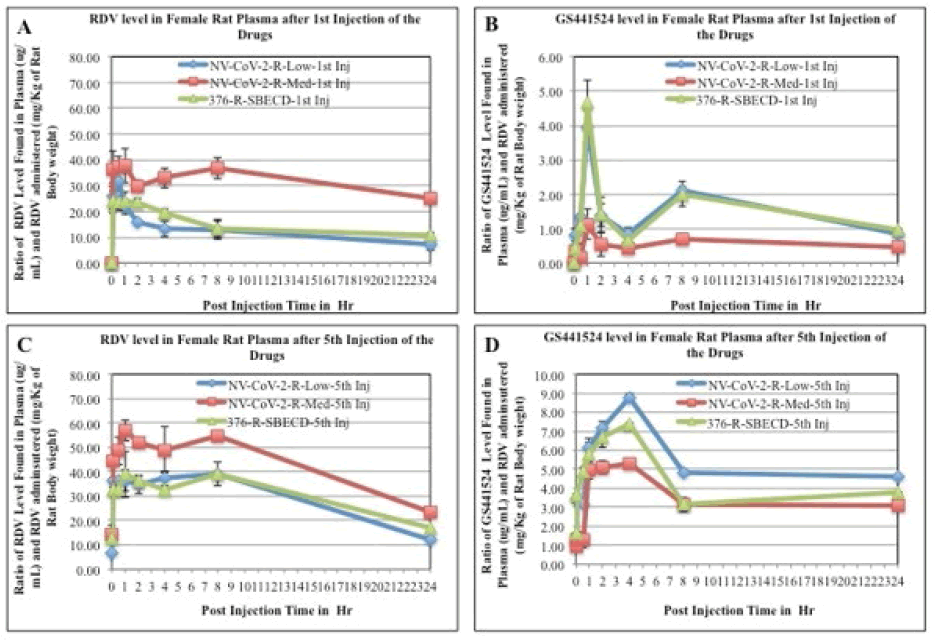
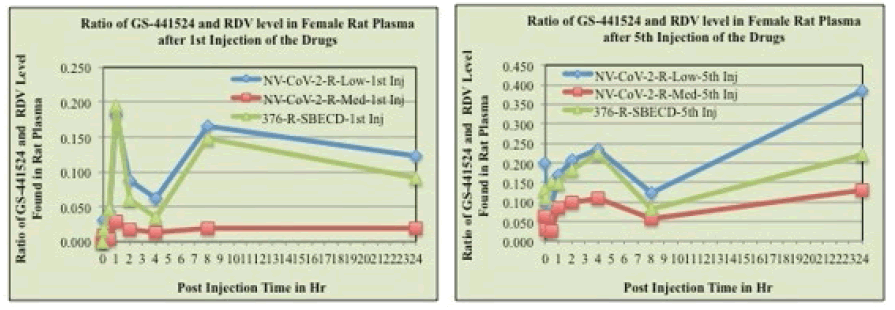
The mean plasma concentration-time profiles of Remdesivir after intravenous administration of Remdesivir in rats were presented in Figure 3, (panel A and C). Immediately after Remdesivir infusion, peak serum concentrations were observed in both male and female rats. Rapid decay of plasma concentration of RDV was observed 1 h after infusion, accompanied by the appearance of GS-441524 in there (Panel B and D). At 24 h after intravenous administration, the concentrations of Remdesivir were below the detection level in rat plasma.
Figure 3: RDV and GS-441524 values in Rat plasma after 1st injection (panels, A, B, respectively) and 5th injections (panels C, D, respectively) of the drugs. The blood samples collected at different time points after drug administration i.v. to the animals were measured for RDV as well as GS-441524 level as described in the method section. The values obtained as μg/mL were normalized by dividing with the RDV amount administered (mg/kg of rat body weight). Each data points are the mean (±SD) of 3 values and the experiment was repeated three times with similar results
Pharmacokinetics of GS-441524
The mean plasma concentration-time profiles of GS441524 after intravenous administration of Remdesivir in rats (female) were presented in Figure 3 (panels, B, D). Data demonstrated that 1 h after Remdesivir infusion a GS- 441524 peak serum concentration was present in rat plasma. In humans, similar rise and fall peaks of GS-445124 plasma concentrations after IV administration RDV were reported [23]. Further, a comparison of the concentration ratio of GS-441524 and RDV (GS-5734) was determined from Figure 3. Results are evident from the Figures that when encapsulated in the polymer, NV-CoV-2, breakdown products level, GS-441524 is less than in the other conditions (Figure 4).
Discussion
Remdesivir is a nucleotide analogue RNA polymerase inhibitor that showed decreased viral load and improved pulmonary function in an animal model of SARSCoV and MERS-CoV infection [3,8]. GS-441524 is the predominant metabolite in the bloodstream when Remdesivir is administered IV [3,4,17]. Hydrolytic enzymes are ubiquitous in serum [15,16,24]. GS441524, also have been shown to inhibit SARS-CoV-2 replication with IC50=0.70 μM compared to Remdesivir (IC50=1.35 μM) in Vero E6 cells [18]. Moreover, GS-441524 showed minimal cytotoxicity in vivo [1,25]. However, GS-441524 is not a better choice over remdesivir, because GS-441524 is the predominant form (74%) of RDV excretion via urine. Plasma concentrations of GS-441524 were found in a patient with renal impairment, indicating that renal excretion was a major route of RDV elimination from the body [21,26]. Further, the cell permeability of GS-441524 is very poor compared to RDV [27-30]. Lastly, RDV is the only drug that is FDA approved, but not the GS-441524.
Conclusion
All studies that have investigated the PK of Remdesivir in Non-Human Primates (NHP) have concluded that intact Remdesivir exhibits a short plasma half-life of about 0.4 h in serum, with “persistence” of the downstream nucleoside, GS-441524. With reference to the above observation we have presented here that the RDV when encapsulated with our BioPolymer NV-CoV-2, the half-life has increased in vivo.
Acknowledgement
We acknowledge all our colleagues, Secretaries for their help during the preparation of the manuscript by providing all the relevant information.
Author contributions
All the authors contributed equally to preparing this article, read and approved the final manuscript.
Ethical Statement
All the animal handling experiments are done by Dr Krishna Menon from AR Biosystems, (17633 Gunn Highway, Odessa, FL 33556), as per the protocol #IACUC No. 14/17ARB. In brief, there are no known acceptable alternatives to the use of live animals to accomplish the purpose of this study. The animal is a standard model for non-clinical studies, for which there is a large historical database, and has been found a suitable model in previous investigations with the same disease models.
This study was designed to use the fewest number of animals possible consistent with the scientific objectives of the study, the contemporary scientific standards, and in consideration of applicable regulatory requirements. The number of animals used is considered the minimum to make meaningful biological comparisons and for statistical calculations.
Conflict of interest
Author, Anil Diwan, was employed by the company Nanoviricides, Inc. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
Fundings from Nanoviricide, Inc.
References
- Siegel D, Hui HC, Doerffler E, et al. Discovery and Synthesis of a Phosphoramidate Prodrug of a Pyrrolo[2,1-f][triazin-4-amino] Adenine C-Nucleoside (GS-5734) for the Treatment of Ebola and Emerging Viruses. J Med Chem. 60:1648-1661 (2017).
Google Scholar Crossref - Schneider B, Xu YW, Sellam O, et al. Pre-steady State of Reaction of Nucleoside Diphosphate Kinase with Anti-HIV Nucleotides. J Biol Chem. 273:11491-11497 (1998).
Google Scholar Crossref - Sheahan TP, Sims AC, Graham RL, et al. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci Transl Med. 9:eaal3653 (2017).
Google Scholar Crossref - Warren TK, Jordan R, Lo MK, et al. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 531:381-385 (2016).
Google Scholar Crossref - Jordan PC, Liu C, Raynaud P, et al. Initiation, extension, and termination of RNA synthesis by a paramyxovirus polymerase. PLoS Pathog. 14:e1006889 (2018).
Google Scholar Crossref - Tchesnokov EP, Feng JY, Porter DP, et al. Mechanism of inhibition of Ebola virus RNAdependent RNA polymerase by remdesivir. Viruses. 11:E326 (2019).
Google Scholar Crossref - Lo MK, Jordan R, Arvey A, et al. GS-5734 and Its Parent Nucleoside Analog Inhibit Filo-, Pneumo-, and Paramyxoviruses. Sci Rep. 7:43395 (2017).
Google Scholar Crossref - Sheahan TP, Sims AC, Leist SR, et al. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat Commun. 11:222 (2020).
Google Scholar Crossref - Wang M, Cao R, Zhang L, et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 30:269-271 (2020).
Google Scholar Crossref - Brown AJ, Won JJ, Graham RL, et al. Broad spectrum antiviral remdesivir inhibits human endemic and zoonotic delta coronaviruses with a highly divergent RNA dependent RNA polymerase. Antivir Res. 169:104541 (2019).
Google Scholar Crossref - de Wit E, Feldmann F, Cronin J, et al. Prophylactic and therapeutic remdesivir (GS-5734) treatment in the rhesus macaque model of MERS-CoV infection. Proc Natl Acad Sci U S A. 117:6771-6776 (2020).
Google Scholar Crossref - Beigel JH, Tomashek KM, Dodd LE, et al. Remdesivir for the Treatment of Covid-19- Final Report. N Engl J Med. 383:1813-1826 (2020).
Google Scholar Crossref - https://www.fda.gov/drugs/news-events-humandrugs/remdesivir-veklury-approvaltreatment-covid-19-evidence-safety-andefficacy
Google Scholar Crossref - Wang Y, Zhou F, Zhang D, et al. Evaluation of the efficacy and safety of intravenous remdesivir in adult patients with severe COVID-19: Study protocol for a phase 3 randomized, double-blind, placebo-controlled, multicentre trial. Trials. 21:422 (2020).
Google Scholar Crossref - Cooke AM, Baron DN. Section of Medicine with Section of Pathology-Serum Enzymes in Clinical Practice. Royal Soc Med. 56:173-177 (1963).
Google Scholar Crossref - Bahar FG, Ohura K, Ogihara T, et al. Species Difference of Esterase Expression and Hydrolase Activity in Plasma. J Pharm Sci. 101:3979-3988 (2012).
Google Scholar Crossref - Williamson BN, Feldmann F, Schwarz B, et al. Clinical benefit of remdesivir in rhesus macaques infected with SARS-CoV-2. Nature. 585:273-276 (2020).
Google Scholar Crossref - Pruijssers AJ, George AS, Schäfer A, et al. Remdesivir Inhibits SARS-CoV-2 in Human Lung Cells and Chimeric SARS-CoV Expressing the SARS-CoV-2 RNA polymerase in Mice. Cell Rep. 32:107940 (2020).
Google Scholar Crossref - Choy KT, Wong AY, Kaewpreedee P, et al. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antiviral Res. 178:104786 (2020).
Google Scholar Crossref - Yan VC, Florian L. Advantages of the Parent Nucleoside GS-441524 over Remdesivir for Covid-19 Treatment. ACS Med Chem Lett. 11:1361-1366 (2020).
Google Scholar Crossref - https://www.ema.europa.eu/en/documents/other/summarycompassionate-use-remdesivirgilead_en.pdf
Google Scholar Crossref - Chakraborty A, Diwan A, Arora V, et al. Nanoviricde platform Technology Based NV-CoV-2 polymer: A safest and an efficient regimen against COVID-19, in vivo. EC Pharmacol Toxicol. 10:1 (2022).
Google Scholar Crossref - Li Y, Cao L, Li G, et al. Remdesivir Metabolite GS-441524 Effectively Inhibits SARS-CoV-2 Infection in Mouse Models. J Med Chem. 65:2785-2793 (2022).
Google Scholar Crossref - Yong JM. Origins of Serum Alkaline Phosphatase. J Clin Pathol. 20:647-653 (1967).
Google Scholar Crossref - Cho A, Saunders OL, Butler T, et al. Synthesis and antiviral activity of a series of 1′-substituted 4-aza-7,9-dideazaadenosine C-nucleosides. Bioorg Med Chem Lett. 22:2705-2707 (2012).
Google Scholar Crossref - Albarello F, Pianura E, Di Stefano F, et al. 2019-novel coronavirus severe adult respiratory distress syndrome in two cases in Italy: An uncommon radiological presentation. Int J Infect Dis. 93:192-197 (2020).
Google Scholar Crossref - De Clercq E. Strategies in the design of antiviral drugs. Nat Rev Drug Discov. 1:13-25 (2002).
Google Scholar Crossref - Mehellou Y, Balzarini J, McGuigan C. Aryloxy phosphoramidate triesters: a technology for delivering monophosphorylated nucleosides and sugars into cells. Chem Med Chem. 4:1779-1791 (2009).
Google Scholar Crossref - Seley-Radtke KL, Yates MK. The evolution of nucleoside analogue antivirals: A review for chemists and non-chemists. Part 1: Early structural modifications to the nucleoside scaffold. Antiviral Res. 154:66-86 (2018).
Google Scholar Crossref - Eastman RT, Roth JS, Brimacombe KR, et al. Remdesivir: A Review of Its Discovery and Development Leading to Emergency Use Authorization for Treatment of COVID-19. ACS Cent Sci. 6:672-683 (2020).
Google Scholar Crossref