Special Report - Imaging in Medicine (2013) Volume 5, Issue 5
Medical imaging for cardiac regeneration using resident cardiac stem cells
Sung Hyun Choi1, So Young Yoo2,3, Sae Mi Yoo1, Kyeong Been Lee2,3, Sang Hong Baek1 and Sang-Mo Kwon*2,31 Laboratory of Cardiovascular Regeneration, Division of Cardiovascular Medicine, Seoul St Mary’s Hospital, The Catholic University of Korea School of Medicine, 505, Banpo-dong, Seocho-gu, Seoul 137-70, South Korea
2Laboratory of Vascular Medicine& Stem Cell Biology, Convergence Stem Cell Research Center, Yangsan, South Korea
3 Immunoregulatory Therapeutics Group in Brain Busan 21 Project, Department of Physiology, Pusan National University School of Medicine, Beomeo-ri, Mulgeum-eup, Yangsan 626-70, South Korea
Abstract
Keywords
MRI; optical imaging; radionuclide imaging; resident progenitor/cardiac stem cells
Myocardial infarction accelerates cardiomyocyte loss as cardiomyocyte apoptosis and necrosis are caused by a lack of nutrients and oxygen supply brought about by ischemic conditions [1]. Although commonly used methods, such as drug treatment, cardiovascular intervention and heart transplantation, can reduce mortality in cardiac disease patients, these therapies cannot regenerate the lost cardiomyocytes [2]. Diverse research has been carried out to overcome limitations in the regeneration of damaged cardiomyocytes. Stem cell-based therapy has been considered a novel therapeutic strategy for cardiomyocyte regeneration since the early 2000s [3]. Stem cells from various sources, including embryonic stem cells (ESCs), induced pluripotent stem cells (iPSCs) and adult stem cells (ASCs) have shown therapeutic potential in the regeneration of cardiomyocytes.
Until now, stem cell therapy for cardiac regeneration has been actively explored using a variety of different cell types. Cardiac stem cells (CSCs) could be considered as an obvious cell source for regeneration of damaged cardiomyocytes. These cells pre-exist in the adult heart, coexpress early cardiac transcription factors and differentiate toward cardiac lineages, which together indicate an ability to improve cardiac regeneration potential compared with other stem cells [4]. A number of experiments have demonstrated that CSCs may regenerate the myocardium by differentiating into cardiomyocytes and vascular lineages (e.g., endothelial cells and vascular smooth muscle cells) [5]. Some clinical trials using CSCs are currently underway [6–8], but the efficiency of engraftment and [2].
the in vivo survival of transplanted stem cells remain challenges that need to be overcome. Therefore, advanced techniques for measuring the efficacy and survival of transplanted stem cells used in clinical applications, including the tracking of injected exogenous CSCs, are required to achieve a better understanding of organ-regeneration mechanisms and the development of stem cell therapy. Imaging approaches not only play important roles in the judgment of clinical availability, including CSC therapy, but also broaden our knowledge regarding stem cell biology [9,10]. There is accumulating evidence that CSCs transplanted into damaged cardiovascular tissues can regenerate, following analyses by medical imaging methods, such as optical imaging, MRI, CT, SPECT and PET [10,11]. However, the tracking of transplanted CSCs in live organs is incredibly difficult and, until now, known imaging analysis methods cannot perfectly trace transplanted exogenous stem cells. Therefore, more advanced medical imaging approaches should be developed for accurate evaluation of implanted CSCs’ effects in vitro and in vivo.
In this article, we present the classification of CSC sources and the most commonly used medical imaging techniques for CSCs for cardiac regeneration. We also discuss the direction of future research for medical imaging of stem cells for cardiac regeneration.
Stem cell sources for cardiac regeneration
Damaged cardiomyocytes have been shown to regenerate when diverse stem cells, including ESCs, iPSCs and ASCs are applied [2,12]. ESCs are an important source of stem cells for cardiomyocyte regeneration and they can differentiate into diverse cellular lineages, including cardiomyocytes, under specific culture conditions. Some experiments have shown that ESC-derived cardiac cells and cardiomyocytes play crucial roles in regenerating damaged cardiomyocytes through interaction with the host heart tissue [13,14]. Despite the positive effects demonstrated by these studies, typically only a few differentiated cardiomyocytes have innate contractile ability [15], and the mechanisms of differentiation into cardiomyocytes have not been completely demonstrated [16]. In addition, ESC transplantation may have ethical implications and undesirable adverse effects such as teratoma development or immune rejections. In view of these disadvantages, clinical trials using ESCs have not yet been conducted.
In 2006, Yamanaka and Takahashi introduced iPSCs, developed from somatic cells, which could differentiate into diverse cellular lineages [17]. iPSCs have rapidly emerged as a novel stem cell source for cardiovascular regeneration, which overcomes the disadvantage of the ethical discussion regarding ESCs. Zhang et al. have shown that human iPSCs can differentiate into functional cardiomyocytes under specific conditions [18]. Nelson and colleagues have reported that transplanted mouse iPSCs could improve left ventricle function in animal models of ischemic heart disease [19]. Although iPSCs or iPSC-derived cells have shown positive effects on cardiovascular regeneration, the efficacy of cardiomyocyte differentiation is extremely low, and these cells have the same limitations as ESCs, such as teratoma generation, when they are transplanted in vivo [20]. Furthermore, the generation of iPSCs, which includes four reverse transcription factors (e.g., Sox2, Klf4, c-Myc and Oct3/4) via retrovirus transfection, makes them inappropriate, in terms of safety, for clinical application in cardiovascular regeneration. However, these limitations have been continuously overcome by the generation of iPSCs without the use of viral vectors and the c-Myc gene, as evidenced by Okita et al. [21] and Nagakawa et al. [22].
Recently, an interesting study reported that fibroblasts and cardiac fibroblasts can be transdifferentiated into cardiomyocyte-like cells using direct reprogramming techniques [23,24]. Despite the fact that direct reprogramming techniques offer novel strategies for cardiovascular regeneration without stem cell transplantation, there are still limitations, in terms of safety, as retrovirus vectors are also used for genetic modulation.
ASC-based therapy is more likely to flourish than other stem cell-based therapies using ESCs or iPSCs, since ASCs (e.g., bone marrow-derived stem cells [BMSCs], myoblasts [MBs], mesenchymal stem cells [MSCs] and CSCs) reside in diverse organs, including the mammalian heart, and these cells can be easily isolated [12].
Owing to the strong resistance of MBs to ischemic conditions and their potential to differentiate into myotubes in tissue, the first study on stem cell therapy for cardiac regeneration was conducted with transplantation of MBs into ischemic heart tissue [25,26]. Some experiments have shown that transplantation with MBs improved left ventricle function [25]. However, transplanted MBs are unable to differentiate into cardiomyocytes [27], and they have also been associated with serious side effects, such as triggering arrhythmias [28].
BMSCs are one of the best-characterized cells for cardiac regeneration. Diverse studies have demonstrated that the reduction in cardiac function in ischemic heart disease could be recovered after transplantation with BMSCs [29–31]. Despite these positive results, the use of BMSCs is still controversial owing to the lack of definite knowledge to determine the core cells. Hematopoietic stem cells [32–34] and endothelial progenitor cells, which are different types of BMSCs, have the potential for cardiac regeneration via their endothelial lineages and differentiation potentials [35,36]. Despite the positive results from these cell therapies, cell differentiation into cardiomyocytes remains debatable [37–39].
In addition, MSCs are an advantageous stem cell therapeutic resource in adults as they can be isolated from diverse tissues, such as bone marrow, adipose tissue and umbilical cord blood. MSCs can also differentiate into diverse cellular lineages including cardiomyocytes, osteocytes, chondrocytes and adipocytes [40–42]. However, these cells showed a lower rate of survival and differentiation into cardiomyocytes in vivo after transplantation [43]. Another disadvantage is that MSCs often show different characteristics depending on their source or donor owing to the lack of characteristic markers.
As described above, CSCs can regenerate cardiomyocyte populations in the adult heart and, thus, various types of CSCs have been identified by diverse research groups [2].
Various types of resident CSCs used for cardiac regeneration
CSCs in adult hearts can be isolated from the myocardium and epicardium by diverse methods, such as conjugation with specific antibodies and vital dye-exclusion methods. CSCs can thus be classified as described below.
Sca-1-positive cells
•Sca-1 is a well-defined cell surface marker expressed on hematopoietic stem cells [44]. Oh et al. suggested that Sca-1-positive cells isolated from the mouse adult heart can differentiate into GATA4 and Nkx2–5-positive cardiomyocytes following treatment with 5-azacytidine [45]. Matsuura et al. reported that mouse Sca-1-positive CSCs can differentiate into beating cardiomyocytes following treatment with oxytocin [46]. Huang et al. recently reported that Sca-1-positive mouse CSCs protect the myocardium via their paracrine effects in ischemic/reperfusion injury models [47]. Furthermore, some groups reported that Sca-1-positive CSCs isolated from the adult human myocardium also improved cardiac function following transplantation [48,49]. Unfortunately, there is no evidence from clinical trials using Sca-1-positive CSCs.
• c-kit-positive cells
c-kit is also a well-defined stem cell surface marker expressed on diverse stem cells, including CSCs. Beltrami and colleagues first identified c-kit-positive CSCs in the adult rat myocardium and showed that c-kit-positive CSC injection improved cardiac functions following differentiation into cardiomyocytes [50]. Another research group reported that c-kit-positive CSCs isolated from the adult human myocardium can improve cardiac function when they are transplanted [51]. Recently reported research showed that treatment with ephrin A1 enhanced migration of c-kit-positive CSCs and improved left ventricle function [52]. More recently, SCIPIO clinical trials first reported clinical trials of CSCs, which have shown that implantation with c-kit-positive human CSCs can improve the cardiac function [6,7].
• Isl-1-positive cells
Isl-1 plays crucial roles in fetal organ development [53]. Laugwitz et al. first identified Isl-1-positive CSCs in the mammalian fetal heart and found that the cells can differentiate into functional cardiomyocytes in vitro [54]. Moretti et al. reported that embryonic-derived Isl-1-positive CSCs can differentiate into cardiomyocytes and vascular lineages [55]. Although there is diverse evidence showing the positive effects of Isl- 1-positive CSCs [56–58], it is still unclear whether these cells can be classified as ASCs since Isl-1 is predominantly expressed in the fetal heart tissue, rather than adult tissue [59]; moreover, thus far, there is no evidence from clinical trials using Isl-1-positive CSCs.
• Cardiac side population cells
Side population cells can be found in diverse organs, including the skeletal muscle, bone marrow and adipose tissue [60,61]. Cardiac side population cells (CSPCs) can also be isolated from the adult myocardium using an exclusion assay with vital dyes (Hoechst 33342 or Rhodamine 123). Martin et al. first identified CSPCs in the developing adult heart, with these cells expressing Abcg2 [60]. Oyama et al. demonstrated that CSPCs express Bcrp-1 and can differentiate into diverse lineages, including cardiomyocytes [62]. More recently, a high frequency of Brcp- 1-positive CSPCs was detected in ischemic human myocardium [63]. In addition, Yoon et al. reported that transplanted CSPCs improved blood perfusion in a hind limb ischemia model [64]. However, the differentiation mechanisms of these cells are yet to be elucidated.
• Cardiosphere-derived stem cells
Messina et al. were the first to demonstrate that CSC populations can form an aggregated spheroid form in culture (i.e., a cardiosphere) [65]. Cardiosphere-derived stem cells (CDSCs) can also differentiate into cardiomyocytes, endothelial cells and vascular smooth muscle cells both in vitro and in vivo [66]. Diverse animal experiments have been conducted and have shown that transplanted CDSCs improve cardiac function [67,68]. Recently, a clinical trial of transplanted human CDSCs (CADUCEUS), carried out by Makkar et al., demonstrated that implantation of human CDSCs improved ventricle wall thickness but did not show ejection fraction improvement [8]. CDSCs may have disadvantages, such as contamination with cardiac fibroblasts, but this disadvantage can be overcome by using stem cell markers.
• Epicardium-derived cells
CSCs exist not only in the adult mammalian myocardium but also in the epicardium. Some of the epicardial cells covering the primitive heart acquire migration abilities by epithelial–mesenchymal transition [69]. Limana et al. demonstrated that c-kit+/CD34+/CD45- populations exist in human and murine epdicardiums and can differentiate into cardiomyocytes [70]. In addition, Zhou et al. reported that Wt1-positive epicardium-derived cells (EPDCs) also differentiate into cardiomyocytes [71]. Furthermore, some evidence suggested that thymosin-b4 improved migration of EDPCs and Wt1-positive EDPC populations primed with thymosin-b4 differentiated into cardiomyocytes [72,73]. However, the differentiation potential of EDPCs is still controversial. Thus, further development of cellular surface markers to isolate EDPCs may be required.
Medical imaging of resident CSCs
As described above, CSC-based cell therapies have shown improvement of cardiac function in animal experiments [74]; however, some clinical evidence has shown that cell-therapeutic effects are rather weak since stem cell therapy has fundamental limitations [6–8], such as inefficient long-term engraftment and low survival rates of stem cells transplanted into target organs [2,12]. To overcome these limitations, stem cell research requires a more suitable and advanced technology for monitoring the state of transplanted stem cells. Medical imaging technology is rapidly developing and could provide wider insights into the understanding of stem cell fate, migration and survival in vivo.
Optical imaging analysis
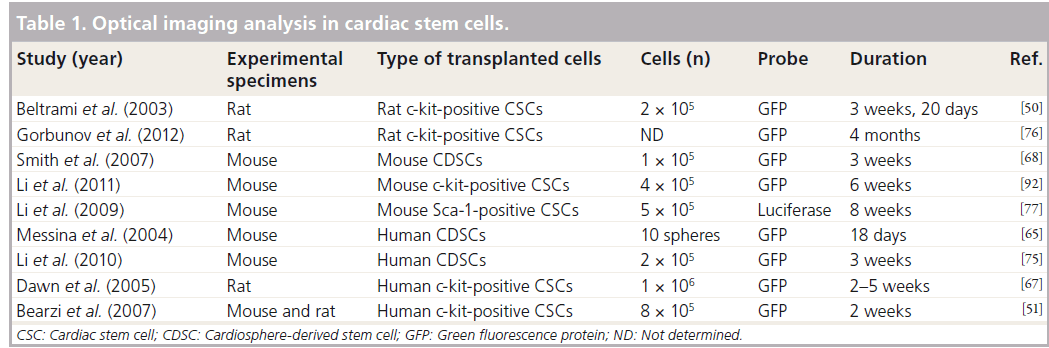
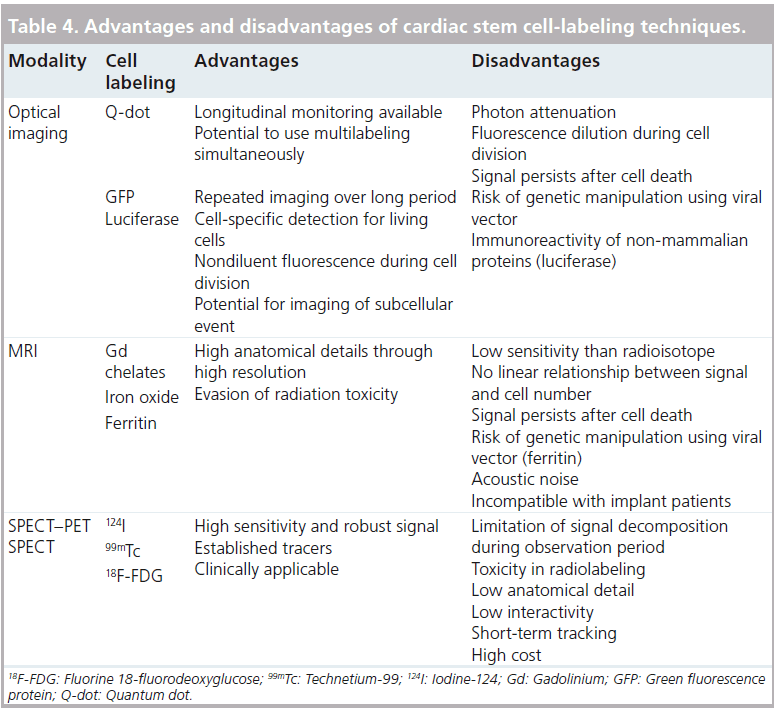
Optical imaging analysis has been widely used in the monitoring and tracking of transplanted stem cells in vivo. To monitor transplanted stem cells, diverse probes, such as chemical (e.g., fluorescence dyes or quantum dots [Q-dots]) or biological reporters (e.g., fluorescence proteins), are used to label the stem cells, and these methods have been previously well demonstrated (Table 1). Smith and colleagues showed that 1 × 105 of Dil-labeled human CDSCs could be traced in mouse-infarcted myocardium even at 3 weeks after their transplantation [68]. Furthermore, Bearzi et al. reported that 8 × 105 of Q-dot- 605-labeled human c-kit-positive CSCs could be detected in mouse- and rat-infarcted myocardium 2 weeks after transplantation [51]. These chemical reporter-labeled techniques are easily applicable to post-transplant stem cell tracking; however, they have some limitations. Fluorescence intensity measured using chemical reporters could be diluted by cell division and detected in vivo regardless of cell survival [10].
The limitations of chemical reporter-labeled stem cell tracking techniques can be overcome using biological reporters such as fluorescence proteins and luciferase. Some evidence has shown that green fluorescent protein-labeled human CDSCs can be detected in infarcted rat myocardium until 3 weeks after transplantation [75]. More recently, Gorbunov and colleagues reported that the homing capability of rat c-kit-positive CSCs was enhanced by resveratrol priming, which could be traced long term (4 months) using a green fluorescent proteinlabeling technique [76]. Furthermore, in 2009, Li et al. reported that mouse Sca-1-positive CSCs could be traced using diverse imaging analysis methods. This study showed that mouse Sca- 1-positive CSCs, labeled using firefly luciferase transfection, were successfully detected 8 weeks after implantation by live imaging [77]. Cells transfected with fluorescent protein gene and luciferase are available for transplanted cell tracking. Most of the accumulating evidence suggests that diverse sources of CSCs have been traced effectively after in vivo transplantation using green fluorescence labeling methods (Table 1). Although stem cell labeling using fluorescence protein has been shown to appropriately overcome the disadvantages of chemical dyes or Q-dots, this technique has disadvantages. Most experiments have shown that transplanted stem cells could be detected after sacrificing the animals using an invasive method, but this limitation could be overcome using a luciferase genebased cell-labeling technique. This technique is needed to appropriately extract the substrate (such as d-luciferin) without sacrificing the animal for cell tracking.
However, these techniques are not suitable for clinical applications, as they are based on genetic manipulation. In addition, the detection of transplanted stem cells using light-based imaging analysis has the fundamental limitation that it can only penetrate tissues with a depth of <10 cm [9].
Furthermore, Goichberg et al. have reported that CSCs could be detected 2 weeks after transplantation using in situ hybridization using the Y chromosome and human-specific Alu sequence. However, these tracking techniques are considered inappropriate for stem cell tracking in clinical trials [52].
•MRI analysis
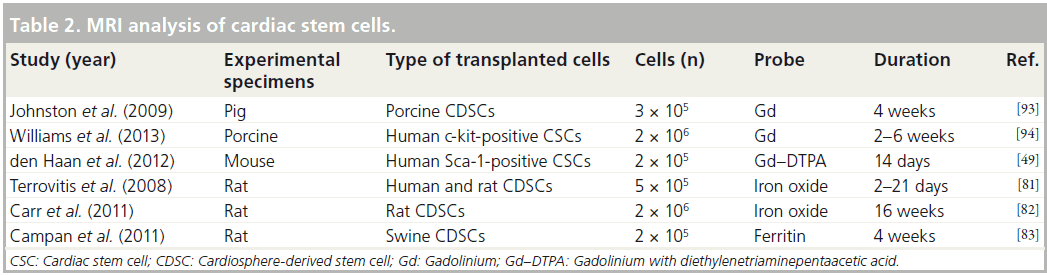
MRI provides excellent resolution of images of cardiac anatomy and function [78]. Furthermore, MRI has benefits for in vivo tracking of stem cells due to its safety and 3D reconstructive capability [79]. Thus, MRI analysis has been widely used to monitor transplanted CSCs and analyze their functions (Table 2).
Diverse MRI analyses with gadolinium contrast reagents have been used extensively as noninvasive tools for the assessment of myocardial infarct size following CSC transplantation in clinical trials. In the SCIPIO clinical trial, implantation of human c-kit-positive CSCs reduced the infarct sizes to 24 and 30% at 4 and 12 months after transplantation, respectively, and myocardial infarct sizes were properly assessed using gadolinium-based MRI in ischemic myopathy patients (SCIPIO clinical trial) [6,7]. Another clinical trial has reported that injected human CDSCs altered ventricle wall thickness, but did not significantly improve ejection fraction [8].
Although gadolinium-based MRI has been used to effectively assess myocardial infarct sizes with excellent resolution after CSC transplantation, intravenous gadolinium contrast reagents are not sufficient to trace transplanted CSCs as these reagents do not provide cell-specific imaging [80].
Recent MRI clinical trials were performed for functional assessment of cardiac performance only; however, some other researchers have demonstrated that transplanted CSCs can be traced by MRI with advanced contrast reagents [6–8]. Terrovitis and colleagues reported that ferumoxide- labeled rat CDSCs can be detected by 3T MRI at 3 weeks post-transplantation, although ferumoxide signaling was also detected in dead CDSCs and macrophages [81]. Furthermore, Carr et al. reported that fluorescent micron-sized particles of iron oxide-labeled rat CDSCs could be detected at 16 weeks post-transplantation [82]. This result strongly indicates that MRI with advanced contrast reagents offers benefits in terms of tracking transplanted CSCs in vivo. However, iron oxide-based MRI reagents can be diluted by subsequent cell divisions, and this remains a limitation of this technique.
Interestingly, recently reported evidence has shown that transplanted stem cells can be traced using the induction of iron-storage proteins, such as ferritin heavy chain. According to this report, porcine CDSCs transfected with human ferritin heavy chain can be detected at 6 weeks post-transplantation in infarcted border zones using 1.5T MRI [83]. This research indicates that transplanted CSCs can be traced using biological MRI reagents, which overcomes the limitation of reagent dilution brought about by CSC division. However, the fact that MRI analysis is incompatible with pacemaker and implantable cardioverters/defibrillators remains a challenge in cardiology [80].
• Radionuclide imaging analysis
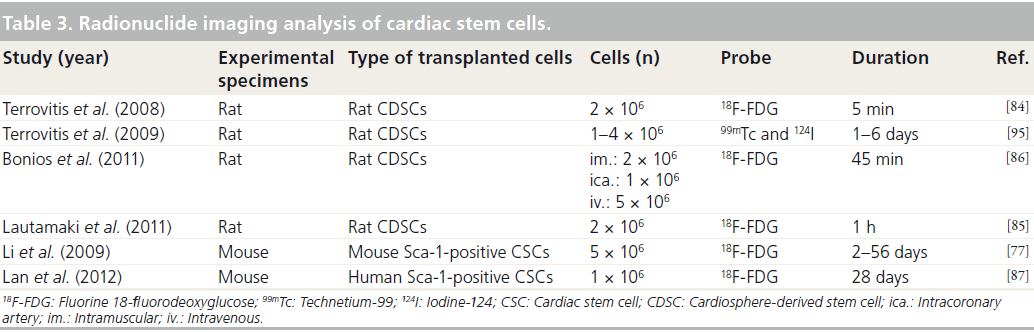
Radionuclide imaging analyses, including PET and SPECT, have high sensitivities in the measurement of cell tracking since the body does not emit inherent signals that interfere with radioprobes [11]. Several positron-emitting probes and radionuclides have been used in preclinical studies to study stem cell migration and engraftment (Table 3).
Terrovitis and their colleagues have reported that implanted rat CDSCs could be traced for shor-term engraftment for 1–6 days post-transplantation by labeling with technetium-99 and iodine-124 [84]. This study showed that technetium-99 and iodine-124 uptake increased using sodium–iodine symporter and did not affect cellular viability and proliferation.
Fluorine 18-fluorodeoxyglucose (18F-FDG) is widely used in PET imaging and diverse research groups have demonstrated cellular tracking and cardiac functional analysis using 18F-FDG-labeled CSCs (Table 3). The shortterm engraftment of transplanted CSCs has previously been measured using an 18F-FDG labeling protocol. Lautamaki et al. have shown that only 20% (400,000 cells) of cells injected into rat myocardium were detected at 1-h post-transplantation, out of the 2 million 18F-FDGlabeled rat CDSCs initially transplanted [85]. Furthermore, Bonios and colleagues measured cellular retention rates following different 18F-FDG-labeled rat CDSC injection protocols, and showed that 12.0, 15.4 and 0.8% of 18F-FDG-labeled CDSCs were detected at 45 min after intramuscular, intracoronary artery and intravenous injections, respectively [86]. These results indicate that radio probe-labeling methods have the advantage of short-term tracking for injected CSCs; however, these methods have limitations in terms of monitoring the long-term engraftment of injected CSCs owing to the short half-life of radio probes. Recently, Lan et al. reported that 18F-FDG-labeled human Sca-1-positive CSCs were detected in the mouse myocardium, but the signaling intensities of PET and bioluminescence imaging (BLI) were not matched at 7 days after transplantation (PET: 44.7 ± 3.2%; BLI: 22.7 ± 11.5%) [87]. In addition, the decrement of signal intensity rates for PET and BLI during transplanted cell tracking were also different (PET: -D31.5%; BLI: -D12.1%). These different results were related to the characteristics of radio probes (short half-life) and bioluminescence (short penetration depth) [87].
Based on the above experiments, the radio probe-based CSC labeling technique is an appropriate method for the evaluation of the short-term proliferation of transplanted CSCs, but this application may have some limitations in terms of monitoring the differentiation of injected CSCs, owing to the short half-life of radio probes. Although long half-life radio probes may be useful for evaluating the differentiation potential of injected CSCs, they may not be appropriate for clinical applications owing to the safety issues related to radiation accumulation. Thus, advanced radio probe-based cell monitoring techniques should be developed for the long-term tracking of injected CSCs.
• Other imaging modalities: echocardiography
Echocardiography is widely used to assess cardiac function and evaluate stem cell therapy efficacy. In addition, echocardiography could be an attractive modality of stem cell tracking owing to the low cost and lack of radiation toxicity. Despite these attractive qualities, echocardiography has some disadvantages, such as low anatomical resolution and low stem cell qualification accuracy. For these reasons, there is not yet evidence of CSC tracking using echocardiography. However, only a few studies have reported tracking of MSCs and EPCs using microbubbles [88,89] and CliniMACS nanoparticles [90,91] by echocardiography.
Conclusion
Previous studies have shown that CSCs exert positive effects on cardiac regeneration, and this has been successfully confirmed by tracking injected CSCs with various medical imaging methods. However, no single imaging analysis is sufficient for tracking injected CSCs as each imaging analysis modality has advantages and disadvantages (Table 4). Therefore, the application of diverse approaches using multiple imaging analyses will provide better insights toward understanding the proliferation, differentiation and migration of injected CSCs in vivo. Furthermore, the development of advanced imaging reagents (to overcome existing efficacy and safety limitations) and multifunctional imaging modalities (multiimaging analysis systems using a single piece of equipment) will lead us toward the next generation of monitoring systems used in the research of transplanted CSCs.
Future perspective
Medical imaging clearly shows benefits in terms of tracking transplanted CSCs in vivo, but the advantages and disadvantages of their clinical application must be clearly distinguished. As described above, the diverse probes used to label CSCs have shown different results in terms of tracking transplanted CSCs in the long or short term. Therefore, appropriate imaging probes should be developed for successfully tracing transplanted CSCs. The primary consideration in the development of imaging probes is that these agents should not generate toxic products or contain contaminants. Furthermore, multifunctional image agents must be developed, as tracking transplanted CSCs is extremely complex and most imaging analysis methods are dependent on a single image modality. One final consideration is that agents should not alter the cellular proliferation, stemness and differentiation capability of labeled CSCs.
Financial & competing interests disclosure
This work was supported by grants from the National Research Foundation funded by the Korean goverment (MEST; 2010- 0020260, 20120618, 2012M3A9C6049720) and also supplerted by the Brain Busan 21 Project in 2013. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
- Diwan A, Krenz M, Syed FM et al. Inhibition of ischemic cardiomyocyte apoptosis through targeted ablation of Bnip3 restrains postinfarction remodeling in mice. J. Clin. Invest. 117(10), 2825–2833 (2007).
- Choi SH, Jung SY, Kwon SM et al. Perspectives on stem cell therapy for cardiac regeneration. Advances and challenges. Circ. J. 76(6), 1307–1312 (2012).
- Abdel-Latif A, Bolli R, Tleyjeh IM et al. Adult bone marrow-derived cells for cardiac repair: a systematic review and metaanalysis. Arch. Intern. Med. 167(10), 989–997 (2007).
- Barile L, Messina E, Giacomello A et al. Endogenous cardiac stem cells. Prog. Cardiovasc. Dis. 50(1), 31–48 (2007).
- Baddour JA, Sousounis K, Tsonis PA. Organ repair and regeneration: an overview. Birth Defects Res. C Embryo Today 96(1), 1–29 (2012).
- Bolli R, Chugh AR, D’Amario D et al. Cardiac stem cells in patients with ischaemic cardiomyopathy (SCIPIO): initial results of a randomised Phase 1 trial. Lancet 378(9806), 1847–1857 (2011).
- Chugh AR, Beache GM, Loughran JH et al. Administration of cardiac stem cells in patients with ischemic cardiomyopathy: the SCIPIO trial: surgical aspects and interim analysis of myocardial function and viability by magnetic resonance. Circulation 126(11 Suppl. 1), S54–S64 (2012).
- Makkar RR, Smith RR, Cheng K et al. Intracoronary cardiosphere-derived cells for heart regeneration after myocardial infarction (CADUCEUS): a prospective, randomised Phase 1 trial. Lancet 379(9819), 895–904 (2012).
- Chemaly ER, Yoneyama R, Frangioni JV et al. Tracking stem cells in the cardiovascular system. Trends Cardiovasc. Med. 15(8), 297–302 (2005).
- Fu Y, Azene N, Xu Y et al. Tracking stem cells for cardiovascular applications in vivo: focus on imaging techniques. Imaging Med. 3(4), 473–486 (2011).
- Chan AT, Abraham MR. SPECT and PET to optimize cardiac stem cell therapy. J. Nucl. Cardiol. 19(1), 118–125 (2012).
- Segers VF, Lee RT. Stem-cell therapy for cardiac disease. Nature 451(7181), 937–942 (2008).
- Xue T, Cho HC, Akar FG et al. Functional integration of electrically active cardiac derivatives from genetically engineered human embryonic stem cells with quiescent recipient ventricular cardiomyocytes: insights into the development of cell-based pacemakers. Circulation 111(1), 11–20 (2005).
- Caspi O, Huber I, Kehat I et al. Transplantation of human embryonic stem cell-derived cardiomyocytes improves myocardial performance in infarcted rat hearts. J. Am. Coll. Cardiol. 50(19), 1884–1893 (2007).
- Mignone JL, Kreutziger KL, Paige SL et al. Cardiogenesis from human embryonic stem cells. Circ. J. 74(12), 2517–2526 (2010).
- Condorelli G, Catalucci D. Human stem cells for heart failure treatment ready for prime time? J. Am. Coll. Cardiol. 50(19), 1894–1895 (2007).
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126(4), 663–676 (2006).
- Zhang J, Wilson GF, Soerens AG et al. Functional cardiomyocytes derived from human induced pluripotent stem cells. Circ. Res. 104(4), e30–e41 (2009).
- Nelson TJ, Martinez-Fernandez A, Yamada S et al. Repair of acute myocardial infarction by human stemness factors induced pluripotent stem cells. Circulation 120(5), 408–416 (2009).
- Yoshida Y, Yamanaka S. iPS cells: a source of cardiac regeneration. J. Mol. Cell. Cardiol. 50(2), 327–332 (2011).
- Okita K, Nakagawa M, Hyenjong H et al. Generation of mouse induced pluripotent stem cells without viral vectors. Science 322(5903), 949–953 (2008).
- Nakagawa M, Koyanagi M, Tanabe K et al. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat. Biotechnol. 26(1), 101–106 (2008).
- Ieda M, Fu JD, Delgado-Olguin P et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 142(3), 375–386 (2010).
- Song K, Nam YJ, Luo X et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature 485(7400), 599–604 (2012).
- Menasche P. Skeletal myoblasts as a therapeutic agent. Prog. Cardiovasc. Dis. 50(1), 7–17 (2007).
- Leor J, Patterson M, Quinoness MJ et al. Transplantation of fetal myocardial tissue into the infarcted myocardium of rat. A potential method for repair of infarcted myocardium? Circulation 94(9 Suppl.), II332–II336 (1996).
- Reinecke H, Poppa V, Murry CE. Skeletal muscle stem cells do not transdifferentiate into cardiomyocytes after cardiac grafting. J. Mol. Cell. Cardiol. 34(2), 241–249 (2002).
- Eisen HJ. Skeletal myoblast transplantation: no MAGIC bullet for ischemic cardiomyopathy. Nat. Clin. Pract. Cardiovasc. Med. 5(9), 520–521 (2008).
- Kajstura J, Rota M, Whang B et al. Bone marrow cells differentiate in cardiac cell lineages after infarction independently of cell fusion. Circ. Res. 96(1), 127–137 (2005).
- Yoon YS, Wecker A, Heyd L et al. Clonally expanded novel multipotent stem cells from human bone marrow regenerate myocardium after myocardial infarction. J. Clin. Invest. 115(2), 326–338 (2005).
- Orlic D, Kajstura J, Chimenti S et al. Mobilized bone marrow cells repair the infarcted heart, improving function and survival. Proc. Natl Acad. Sci. USA 98(18), 10344–10349 (2001).
- Mansour S, Roy DC, Bouchard V et al. COMPARE-AMI trial: comparison of intracoronary injection of CD133+ bone marrow stem cells to placebo in patients after acute myocardial infarction and left ventricular dysfunction: study rationale and design. J. Cardiovasc. Transl. Res. 3(2), 153–159 (2010).
- Orlic D, Kajstura J, Chimenti S et al. Bone marrow cells regenerate infarcted myocardium. Nature 410(6829), 701–705 (2001).
- Templin C, Kotlarz D, Faulhaber J et al. Ex vivo expanded hematopoietic progenitor cells improve cardiac function after myocardial infarction: role of beta-catenin transduction and cell dose. J. Mol. Cell. Cardiol. 45(3), 394–403 (2008).
- Jujo K, Ii M, Losordo DW. Endothelial progenitor cells in neovascularization of infarcted myocardium. J. Mol. Cell. Cardiol. 45(4), 530–544 (2008).
- Leone AM, Rutella S, Giannico MB et al. Effect of intensive vs standard statin therapy on endothelial progenitor cells and left ventricular function in patients with acute myocardial infarction: statins for regeneration after acute myocardial infarction and PCI (STRAP) trial. Int. J. Cardiol. 130(3), 457–462 (2008).
- Balsam LB, Wagers AJ, Christensen JL et al. Haematopoietic stem cells adopt mature haematopoietic fates in ischaemic myocardium. Nature 428(6983), 668–673 (2004).
- Gruh I, Beilner J, Blomer U et al. No evidence of transdifferentiation of human endothelial progenitor cells into cardiomyocytes after coculture with neonatal rat cardiomyocytes. Circulation 113(10), 1326–1334 (2006).
- Murry CE, Soonpaa MH, Reinecke H et al. Haematopoietic stem cells do not transdifferentiate into cardiac myocytes in myocardial infarcts. Nature 428(6983), 664–668 (2004).
- Barry FP, Murphy JM. Mesenchymal stem cells: clinical applications and biological characterization. Int. J. Biochem. Cell. Biol. 36(4), 568–584 (2004).
- Xu H, Yang YJ, Qian HY et al. Rosuvastatin treatment activates JAK–STAT pathway and increases efficacy of allogeneic mesenchymal stem cell transplantation in infarcted hearts. Circ. J. 75(6), 1476–1485 (2011).
- Xu X, Xu Z, Xu Y, Cui G. Selective downregulation of extracellular matrix gene expression by bone marrow derived stem cell transplantation into infarcted myocardium. Circ. J. 69(10), 1275–1283 (2005).
- Amado LC, Saliaris AP, Schuleri KH et al. Cardiac repair with intramyocardial injection of allogeneic mesenchymal stem cells after myocardial infarction. Proc. Natl Acad. Sci. USA 102(32), 11474–11479 (2005).
- van de Rijn M, Heimfeld S, Spangrude GJ et al. Mouse hematopoietic stem-cell antigen Sca-1 is a member of the Ly-6 antigen family. Proc. Natl Acad. Sci. USA 86(12), 4634–4638 (1989).
- Oh H, Bradfute SB, Gallardo TD et al. Cardiac progenitor cells from adult myocardium: homing, differentiation, and fusion after infarction. Proc. Natl Acad. Sci. USA 100(21), 12313–12318 (2003).
- Matsuura K, Nagai T, Nishigaki N et al. Adult cardiac Sca-1-positive cells differentiate into beating cardiomyocytes. J. Biol. Chem. 279(12), 11384–11391 (2004).
- Huang C, Gu H, Yu Q et al. Sca-1+ cardiac stem cells mediate acute cardioprotection via paracrine factor SDF-1 following myocardial ischemia/reperfusion. PLoS ONE 6(12), e29246 (2011).
- Smits AM, van Laake LW, den Ouden K et al. Human cardiomyocyte progenitor cell transplantation preserves long-term function of the infarcted mouse myocardium. Cardiovasc. Res. 83(3), 527–535 (2009).
- den Haan MC, Grauss RW, Smits AM et al. Cardiomyogenic differentiation-independent improvement of cardiac function by human cardiomyocyte progenitor cell injection in ischaemic mouse hearts. J. Cell. Mol. Med. 16(7), 1508–1521 (2012).
- Beltrami AP, Barlucchi L, Torella D et al. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell 114(6), 763–776 (2003).
- Bearzi C, Rota M, Hosoda T et al. Human cardiac stem cells. Proc. Natl Acad. Sci. USA 104(35), 14068–14073 (2007).
- Goichberg P, Bai Y, D’Amario D et al. The ephrin A1–EphA2 system promotes cardiac stem cell migration after infarction. Circ. Res. 108(9), 1071–1083 (2011).
- Cai CL, Liang X, Shi Y et al. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev. Cell. 5(6), 877–889 (2003).
- Laugwitz KL, Moretti A, Lam J et al. Postnatal isl1+ cardioblasts enter fully differentiated cardiomyocyte lineages. Nature 433(7026), 647–653 (2005).
- Moretti A, Caron L, Nakano A et al. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell 127(6), 1151–1165 (2006).
- Chien KR, Domian IJ, Parker KK. Cardiogenesis and the complex biology of regenerative cardiovascular medicine. Science 322(5907), 1494–1497 (2008).
- Martin-Puig S, Wang Z, Chien KR. Lives of a heart cell: tracing the origins of cardiac progenitors. Cell Stem Cell 2(4), 320–331 (2008).
- Wu SM, Chien KR, Mummery C. Origins and fates of cardiovascular progenitor cells. Cell 132(4), 537–543 (2008).
- Simpson DL, Mishra R, Sharma S et al. A strong regenerative ability of cardiac stem cells derived from neonatal hearts. Circulation 126(11 Suppl. 1), S46–S53 (2012).
- Martin CM, Meeson AP, Robertson SM et al. Persistent expression of the ATP binding cassette transporter, Abcg2, identifies cardiac SP cells in the developing and adult heart. Dev. Biol. 265(1), 262–275 (2004).
- Challen GA, Little MH. A side order of stem cells: the SP phenotype. Stem Cells 24(1), 3–12 (2006).
- Oyama T, Nagai T, Wada H et al. Cardiac side population cells have a potential to migrate and differentiate into cardiomyocytes in vitro and in vivo. J. Cell Biol. 176(3), 329–341 (2007).
- Emmert MY, Emmert LS, Martens A et al. Higher frequencies of BCRP+ cardiac resident cells in ischaemic human myocardium. Eur. Heart J. doi:10.1093/eurheartj/ehs156 (2012) (Epub ahead of print).
- Yoon J, Choi SC, Park CY et al. Cardiac side population cells exhibit endothelial differentiation potential. Exp. Mol. Med. 39(5), 653–662 (2007).
- Messina E, De Angelis L, Frati G et al. Isolation and expansion of adult cardiac stem cells from human and murine heart. Circ. Res. 95(9), 911–921 (2004).
- Takehara N, Tsutsumi Y, Tateishi K et al. Controlled delivery of basic fibroblast growth factor promotes human cardiosphere-derived cell engraftment to enhance cardiac repair for chronic myocardial infarction. J. Am. Coll. Cardiol. 52(23), 1858–1865 (2008).
- Dawn B, Stein AB, Urbanek K et al. Cardiac stem cells delivered intravascularly traverse the vessel barrier, regenerate infarcted myocardium, and improve cardiac function. Proc. Natl Acad. Sci. USA 102(10), 3766–3771 (2005).
- Smith RR, Barile L, Cho HC et al. Regenerative potential of cardiosphere-derived cells expanded from percutaneous endomyocardial biopsy specimens. Circulation 115(7), 896–908 (2007).
- Gittenberger-de Groot AC, Vrancken Peeters MP, Mentink MM, Gourdie RG, Poelmann RE. Epicardium-derived cells contribute a novel population to the myocardial wall and the atrioventricular cushions. Circ. Res. 82(10), 1043–1052 (1998).
- Limana F, Zacheo A, Mocini D et al. Identification of myocardial and vascular precursor cells in human and mouse epicardium. Circ. Res. 101(12), 1255–1265 (2007).
- Zhou B, Ma Q, Rajagopal S et al. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature 454(7200), 109–113 (2008).
- Smart N, Risebro CA, Melville AA et al. Thymosin beta4 induces adult epicardial progenitor mobilization and neovascularization. Nature 445(7124), 177–182 (2007).
- Smart N, Bollini S, Dube KN et al. De novo cardiomyocytes from within the activated adult heart after injury. Nature 474(7353), 640–644 (2011).
- Chamuleau SA, Vrijsen KR, Rokosh DG et al. Cell therapy for ischaemic heart disease: focus on the role of resident cardiac stem cells. Neth. Heart J. 17(5), 199–207 (2009).
- Li TS, Cheng K, Lee ST et al. Cardiospheres recapitulate a niche-like microenvironment rich in stemness and cell-matrix interactions, rationalizing their enhanced functional potency for myocardial repair. Stem Cells 28(11), 2088–2098 (2010).
- Gorbunov N, Petrovski G, Gurusamy N et al. Regeneration of infarcted myocardium with resveratrol-modified cardiac stem cells. J. Cell. Mol. Med. 16(1), 174–184 (2012).
- Li Z, Lee A, Huang M et al. Imaging survival and function of transplanted cardiac resident stem cells. J. Am. Coll. Cardiol. 53(14), 1229–1240 (2009).
- Kim RJ, Fieno DS, Parrish TB et al. Relationship of MRI delayed contrast enhancement to irreversible injury, infarct age, and contractile function. Circulation 100(19), 1992–2002 (1999).
- Bulte JW, Douglas T, Witwer B et al. Magnetodendrimers allow endosomal magnetic labeling and in vivo tracking of stem cells. Nat. Biotechnol. 19(12), 1141–1147 (2001).
- Lau JF, Anderson SA, Adler E et al. Imaging approaches for the study of cell-based cardiac therapies. Nat. Rev. Cardiol. 7(2), 97–105 (2010).
- Terrovitis J, Stuber M, Youssef A et al. Magnetic resonance imaging overestimates ferumoxide-labeled stem cell survival after transplantation in the heart. Circulation 117(12), 1555–1562 (2008).
- Carr CA, Stuckey DJ, Tan JJ et al. Cardiosphere-derived cells improve function in the infarcted rat heart for at least 16 weeks – an MRI study. PLoS ONE 6(10), e25669 (2011).
- Campan M, Lionetti V, Aquaro GD et al. Ferritin as a reporter gene for in vivo tracking of stem cells by 1.5-T cardiac MRI in a rat model of myocardial infarction. Am. J. Physiol. Heart Circ. Physiol. 300(6), H2238–H2250 (2011).
- Terrovitis J, Kwok KF, Lautamaki R et al. Ectopic expression of the sodium-iodide symporter enables imaging of transplanted cardiac stem cells in vivo by single-photon emission computed tomography or positron emission tomography. J. Am. Coll. Cardiol. 52(20), 1652–1660 (2008).
- Lautamaki R, Terrovitis J, Bonios M et al. Perfusion defect size predicts engraftment but not early retention of intra-myocardially injected cardiosphere-derived cells after acute myocardial infarction. Basic Res. Cardiol. 106(6), 1379–1386 (2011).
- Bonios M, Terrovitis J, Chang CY et al. Myocardial substrate and route of administration determine acute cardiac retention and lung bio-distribution of cardiosphere-derived cells. J. Nucl. Cardiol. 18(3), 443–450 (2011).
- Lan F, Liu J, Narsinh KH et al. Safe genetic modification of cardiac stem cells using a site-specific integration technique. Circulation 126(11 Suppl. 1), S20–S28 (2012). 88 Fujii H, Li SH, Wu J et al. Repeated and
- targeted transfer of angiogenic plasmids into the infarcted rat heart via ultrasound targeted microbubble destruction enhances cardiac repair. Eur. Heart J. 32(16), 2075–2084 (2011).
- Zhong S, Shu S, Wang Z et al. Enhanced homing of mesenchymal stem cells to the ischemic myocardium by ultrasound-targeted microbubble destruction. Ultrasonics 52(2), 281–286 (2012).
- Bara C, Ghodsizad A, Niehaus M et al. In vivo echocardiographic imaging of transplanted human adult stem cells in the myocardium labeled with clinically applicable CliniMACS nanoparticles. J. Am. Soc. Echocardiogr. 19(5), 563–568 (2006).
- Ruhparwar A, Ghodsizad A, Niehaus M et al. Clinically applicable 7-Tesla magnetic resonance visualization of transplanted human adult stem cells labeled with CliniMACS nanoparticles. Thorac. Cardiovasc. Surg. 54(7), 447–451 (2006).
- Li Q, Guo Y, Ou Q et al. Intracoronary administration of cardiac stem cells in mice: a new, improved technique for cell therapy in murine models. Basic Res. Cardiol. 106(5), 849–864 (2011).
- Johnston PV, Sasano T, Mills K et al. Engraftment, differentiation, and functional benefits of autologous cardiosphere-derived cells in porcine ischemic cardiomyopathy. Circulation 120(12), 1075–1083 (2009).
- Williams AR, Hatzistergos KE, Addicott B et al. Enhanced effect of combining human cardiac stem cells and bone marrow mesenchymal stem cells to reduce infarct size and to restore cardiac function after myocardial infarction. Circulation 127(2), 213–223 (2013).
- Terrovitis J, Lautamaki R, Bonios M et al. Noninvasive quantification and optimization of acute cell retention by in vivo positron emission tomography after intramyocardial cardiac-derived stem cell delivery. J. Am. Coll. Cardiol. 54(17), 1619–1626 (2009).