Review Article - Neuropsychiatry (2018) Volume 8, Issue 3
Neurology in the Light of Genomics: Application of NGS and GWAS in Understanding Complex Neurological Disorders
- Corresponding Author:
- Amit Kumar Banerjee
Biology Division, CSIR-Indian Institute of Chemical Technology, Uppal Road, Tarnaka, Hyderabad-500007,Telangana, India
Tel: +91-9885774127
Abstract
Understanding structure and function of human brain remains the most intriguing region of advanced research. Neurological disorders also constitute a major proportion of ongoing brain associated research as several disorders are still not known in a comprehensive manner from a molecular, causative and functional perspective. Earlier linkage studies were considered the most reliable means to answer certain neurogenetics related questions in relevance to heredity and disease inheritance. Technical limitations and ethical issues always hindered the path of deriving more useful insight and understanding neurological diseases such as Schizophrenia, Epilepsy, Parkinson’s disease, Alzheimer’s disease, Bipolar Disorder, ADHD, ALS and other neurological issues. Improvement in the interdisciplinary approaches including genomics based analyses such as NGS, GWAS, GCTA enabled us to explore and understand the causes and factors responsible for onset of neurological conditions and effect of such neurological conditions on human wellbeing. This review summarizes the recent and on-going research carried out globally to unravel the mysteries of various neurological disorders employing latest genomics based technologies. The technological background and disease specific implementations are described in detail.
Keywords
Neurological disorders, NGS, GWAS, GCTA, Schizophrenia, Epilepsy, Parkinson’s disease, Alzheimer’s disease, Bipolar disorder, ADHD, ALS
Introduction
The path of human evolution has been complex and intriguing. As we are being considered the most intelligent and civilized living form on Earth, we need to understand ourselves before anything else. Advances in scientific knowledge have provided us several tools to analyze and understand complex phenomena and unanswered questions related to life.
Human neurology and psychology are such interesting arenas of science where several issues are still either under hypothetical beliefs or not well understood so far [1]. Peeking into this most complex and fascinating subject remained difficult due to various technical, ethical and other limitations. Moreover, apart from the anatomical and physiological aspects, most of the psychological and behavioral understanding remained intangible due to lack of proper experimental evidences. Experimental molecular neurology and psychology or behavioral traits are analogous to the computer hardware and software. It is well evident that a computer successfully operates with the coordinated function of hardware and software, yet it is cumbersome to understand the interaction of hardware and software in real time situation in qualitative or quantitative manner just akin to experimental neurology and psychology.
The scientific fraternity has started to reap the results of the efforts of the Human genome sequencing in many ways. Recent studies suggest that there is a possibility where genome or genetic level studies of humans may help us in understanding the neurology and Psychology and behavioral traits following a proper scientific approach. Even at the genetic level, several hypotheses cannot be proved as any two genomes of the same species also are not exactly same and sample related issues may arise too. There is a hope of connecting the dots between neurology and psychology similar to an object and their shadow following a comprehensive and logical scientific approach.
Burden of Mental Disorders, Care and Prevention
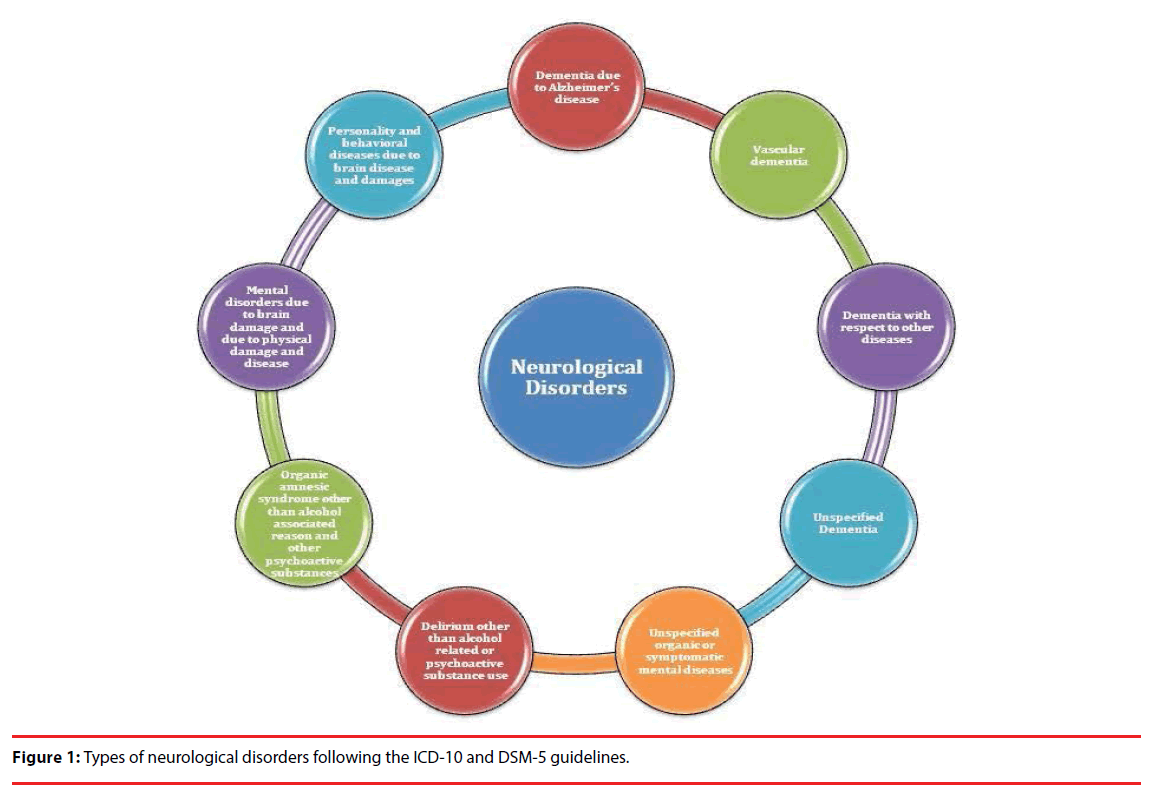
Out of different types of diseases, neurological diseases are on the rise due to the accumulated impact of genetic, environmental and social factors. Mental disorders such as dementia, epilepsy, headache associated disorders, multiple sclerosis, Parkinson disease, Alzheimer’s disease, ADHD, ICD, different neuroinfections, neurological issues related to malnutrition, neuropathic pain, stroke, and traumatic injury related conditions are some of the neurological conditions affecting mankind. According to WHO estimates about the share of DALY (Disability Adjusted Life Years), neurological diseases occupy the top position followed by tuberculosis, HIV, malignant neoplasms and other diseases. Further, the global DALY projection suggests possible increase in Alzheimer and other dementia, cerebrovascular diseases and migraine. Economical group wise, WHO survey and projection suggested that population of high and lower middle income group countries will be among the most affected by one or the other neurological diseases. Geographically, Africa (Afr-D), Americas (Amr-D), Asia (SEAR-D), Europe (Eur-C>Eur-B>Eur-A) and Western Pacific (Wpr-A and Wpr-B) are the regions bearing the brunt of neurological diseases with more than 1200 DALYs per 100000 population [2]. Figure 1 represents the broad types of neurological disorders. Global trends in searching specific neurological problems are represented in Figure 2.
All these neurology associated broad spectrum disease type as per the ICD-10 guidelines (Figure 1) is having one or the other important and neurological diseases which are predominant all over the world in different populations. Moreover, information is available for several neurological conditions in the reputed periodicals as well as in the World Wide Web (Figure 2). We have attempted to report specific information for some of the most common and reported neurological conditions based on their reporting in available journals as well as online media. As this article focuses on the information on application of specific stateof- the-art genomics techniques for neurological diseases, therefore, special care was taken on the availability of such information for a particular condition during short listing of the neurological diseases.
WHO has provided a defined guideline towards prevention of the neurological diseases. According to this protocol, prevention of neurological conditions can be achieved in three stages: primary, secondary and tertiary stage of prevention. At the primary disease prevention stage, treatment measures should be taken prior to the disease onset and required vaccination. During secondary prevention measure, accurate and precise diagnosis, proper treatment plan, assessment of risk factors and prediction of disease progression and compliances should be considered seriously. Use of first line drugs such as antiepileptic and careful patient management as in case of strokes may aid in better health management at community level and save more lives. Tertiary neurological disease management is serious and crucial which actively involves palliative care, rehabilitation, education for the patient’s family and caregivers, avoiding discrimination for patients and race. Complete and comprehensive active system involving medical practitioner, patient, and caregivers can provide remedy and comfort to the patient.
Classification of Neurological Disorders: An Unsolved Puzzle
Neurological disorders are unsolved and complex puzzles which claim maximum scientific and attention with the growing number of cases every year. Interestingly, several diseases are not even identified as a disorder, but increase in number of reports calls for further clinical and molecular investigations. Considering the hurdles and difficulties in diagnosis of neurological diseases, the Diagnostic and Statistical Manual of Mental Disorders (DSM-5) provides a plausible framework and guideline to diagnose neuro-cognitive diseases in agreement with International Classification of Diseases (ICD) 10th edition [3]. As per the ICD-10 guidelines, mental disorders are categorized majorly based on Dementia due to Alzheimer’s disease, vascular dementia, dementia with respect to other diseases, unspecified dementia, organic amnesic syndrome other than alcohol associated reason and other psychoactive substances, delirium other than alcohol related or psychoactive substance use, mental disorders due to brain damage and due to physical damage and disease, personality and behavioral diseases due to brain disease and damages, and unspecified organic or symptomatic mental diseases. Several mental disease conditions are still not specified either due to fuzzy symptoms, disease appearance, expression and progression. Similarly, several neurological conditions are so complex and overlapping in nature that finding a proper and specific cure is tedious.
Genomic Psychology and Cognitive Genomics
Application of genomics and associated technologies has shown remarkable progress in enhancing present understanding of biology. Comparative genomics have matured from the one-gene-one-protein type experiments and analysis to system biology. Interdisciplinary inputs from experimental and theoretical approaches and management of mammoth proportion of data have begun to yield results towards understanding complex and correlated biological phenomenon. Non-invasive techniques such as neuroimaging and genomics are emerging as the real game-changer in enriching our present knowledge. Moreover, the issue of repetitive samples may be resolved in relation to genome based psychological studies using twins as shown by Plomin [1]. The interrelation of psychology and biology is interesting and intriguing at the same time. Both the subjects require bridging the gaps to have a better understanding about the complex traits and associated genetic and environmental conditions.
Genes, Genomes and Intelligence: An Intriguing Paradox
An ever intriguing question is whether intelligence is inheritable? Can we assess the inheritance or at least quantify or predict intelligence from the available genetic information? Intelligence is referred as one of the most precious, complex behavioral trait which actually determines the general life trajectory of an individual. Intelligence involves several indices such as mental and physical health conditions, education, occupation, reaction and response to the surrounding environment and survival capacity etc. [4]. Considering these factors, if we agree that particular genetic traits are involved in expressing complete intelligence in an individual, uncertainty remains regarding the complete expression of those genes associated. Proper phenotypical characteristics may express at any stage of life from infancy to adulthood depending on several cellular and environmental conditions. Therefore, based on the genetic factors, determination of exact quotient is not only cumbersome but next to impossible. There is no single identifier for intelligence; rather it is considered as a collective behavioral expression. Spearman in 1904 defined certain cognitive ability [5] in relevance to intelligence which is used as the rule of thumb to estimate intelligence.
The dogma still lies with the fact that intelligence is inheritable. On the other hand, there are evidences that intelligence cannot be inherited completely. This suggests that interplay of the genetic and environmental factors determines the grade of intelligence in any species. Aggregated small impact of several genes causes the great behavioral alterations phenotypically. But identifying and quantifying those small genetic impacts imparted by several genes is technically labor intensive and cumbersome and not conclusive at the moment.
Moreover, as mentioned earlier, there is no certainty on the expression of particular genes or modification of apparent behavioral traits of an individual. Alteration and modification in gene expression may happen at any stage of life. There are several stimulating questions that exist at present in this regard. It remains to be seen whether the available quantitative genetics based methods can provide any clue or are sufficiently able to quantify all these qualitative factors.
Even if these methods can at least mark and enumerate those genes and their association in relation to inheritability and intelligence, this may result in a tangible solution to the diagnosis and understanding of many neurological problems.
Foreseeing the future from the genomic map has been one of the greatest human desires in this era of genomic revolution.
Genome-wide polygenic score (GPS) based quantitative analyses have been performed to understand plausible relation between DNA and academic achievements [6].
Traditional Molecular Genetics Based Efforts and Other Approaches
Since long, various molecular approaches have been implemented for deciphering the roles of diverse genes in neurological disorders. Traditional chromosomal analysis through karyotyping using cytogenetic methods helped in associating genetics with several diseases such as Down syndrome, Turner syndrome, Wolf-Hirschhorn syndrome and Jacobsen syndrome etc. Prior to the advanced sequencing based technologies, genetic linkage analysis, linkage disequilibrium analysis and association, mutation detection and site directed mutagenesis, implementation of specific animal models were the options available for understanding the molecular aspects of complex neurological disorders. So far, linkage studies are the most accurate method to precisely identify the linkage of physical loci in vicinity or link with a specific disease trait or marker gene. The statistical measure is conducted through logarithms of the odd scores or lod scores which is represented as Z score in the calculation. The genetic linkage technique is being implemented for single gene related disease as well as for polygenic diseases. Linkage association is implemented with reference to the predicted outcome while considering relation of a specific allele and a particular trait including disease trait. Similarly, linkage disequilibrium refers to the presence of specific allele in two different loci with reference to the predicted outcome. Certain association based measures helped in the discovery of the association of apolipoprotein E (ApoE) alleles with Alzheimer’s disease [7]. Similarly, association of tau alleles was found with progressive supranuclear palsy [8]. Spinocerebellar ataxia type 1 was the first neurological disease which was connected to specific features of chromosome [9,10]. Similarly, prior to this outcome, CMT1B was found to be associated. Before the initiation of large cohort based studies at a population level, molecular genetic testing through direct mutation detection or through mutation detection through genetic marker based approaches were widely implemented [11]. After conventional cytogenetic studies, molecular approaches including animal models involving foreign gene expression, gene targeting through knock-out and knock-in methods aided in gaining in-depth insight into the neurological disease conditions. Requirement of better approaches at the population level studies with more quantitative and qualitative genetic information is the need of the time for understanding complex neurological disease conditions.
Advanced Quantitative Techniques in Neurological Disorders
Current scientific effort in this direction is showing promise where experimental and stochastic measures have improved a lot to capture and quantify the small genetic impacts that might be responsible for a large change at the phenotypic trait level. Latest methodologies, such as twin studies and Genome-wide Complex Trait Analysis (GCTA) are such quantitative measurement options [12]. The developer of the GCTA referred it as Genomic-Relatedness- Matrix Restricted Maximum Likelihood (GREML). This method relies on estimating heritability based on the SNP data generated from the advanced sequencing techniques such as Next Generation Sequencing (NGS) protocols.
Next Generation Sequencing (NGS)
Human genome consists of almost 3 billion base pairs which conceal all the secrets of human life. Human Genome Project (HGP) [13] was the first scientific aspiration in the direction of unveiling the mystery of life through sequencing these billions of base-pairs. The whole scientific fraternity held its breath with hopes and anxiety towards the success of the project as well as to witness the outcome of such a gigantic effort in this project. Since then, sequencing technologies have witnessed a revolution. In the last 20 years, there has been a major leap towards sequencing huge number of base-pairs with comparatively low error rate during assembling. Variation in high throughput technology also offered multiple solutions to the technical problems which remained challenging at times. After preparation of the initial draft and publishing the details obtained, further investigations were conducted to fill the lacunae in understanding the overlapping reads, repeat regions and other complex genomic regions. The perception was that it would be simple and linear to unveil the mystery of the human DNA. But it turned out to be more complicated and complex organization in contrary to the initial beliefs.” to “But it turned out to be more complicated and complex in organization in contrary to the initial beliefs. Several new perceptions in relation to the functional aspect of DNA, worthy or “junk” regions emerged after the genome project. Numerous outstanding novel concepts including miRNA (microRNA), siRNA (short interfering RNA), Riboswitch and recently cas (CRISPRassociated system)/CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) based mechanisms came into the lime light due to the ample genome based exercises conducted all over the world in different organisms. The advancement of the NGS technologies contributed immensely to achieve several milestones in genomics. Technologies developed by Ion Torrent, Pacific Biosciences and Illumina aided in revolutionizing the pace of sequencing while maintaining decent amount of accuracy [14,15]. There are basic technical differences among these sequencing machines. The Ion Torrent Personal Genome Machine (PGM) is based on the detection of specific protons generated due to the incorporation of nucleotides during fragmented sequence strand synthesis reaction with the help of Ion Sphere Particles (ISP) [14,15]. The PacBio technology is supported by single molecule real time (SMRT) and zero-mode waveguides (ZMWs) for precisely detecting the fluorescence during the DNA synthesis [14,15]. The Illumina Genome Analyzer follows a principle of synthesis during sequencing methodology [14,15]. The overall objective of all these techniques is to increase the turn-around time with high level of accuracy in terms of genome coverage, GC bias, error rates, SNP calling etc. Parallel processing of multiple samples, inclusion of upfront and library related QC, including advanced library preparation methodology such as Nextera method and enzymatic fragmentation may further aid in improving the output of the system. Table 1 describes some of the NGS based technology platforms and the relevant information.
| Methodology | Company/Organization | Read length | Accuracy |
|---|---|---|---|
| Massively parallel signature sequencing (MPSS) | Lynx Therapeutics | 17-20 bp | - |
| Polony sequencing | Harvard University/Agencourt Biosciences | 26 bp | 99% |
| 454 pyrosequencing | 454 Life Sciences/Roche Diagnostics | 700 bp | 99.9% |
| Illumina (Solexa) sequencing (MiSeq) | Solexa/Illumina | 2 × 300 bp | 89.7% |
| SOLiD sequencing | Applied Biosystems/Life Technologies | 2 × 60 bp reads | 99.85% |
| Ion Torrent semiconductor sequencing | Ion Torrent Systems Inc./Life Technologies | 600 bp | 98% |
| DNA nanoball sequencing | Complete Genomics | 400-500 bp | 1 false variant call per 100 kb |
| Heliscope single molecule sequencing | Helicos Biosciences | 100-200 bp | 99% |
| Single molecule real time (SMRT) sequencing | Pacific Biosciences | 14000 bp | 87% |
| Nanopore DNA sequencing | Agilent Laboratories | 500 kb (library dependent) | 99% |
Table 1: Different NGS methodologies and associated information.
Several novel and upgraded versions of NGS based technology are under development (Table 2), which may further help us in understanding scientific aspects through a comparative genomics approach.
| Methodology (Under Development) | Organization/Collaboration |
|---|---|
| Tunnelling currents DNA sequencing | Nanoelectronics Collaborative Research Center, IIS, The University of Tokyo/University of California/Osaka University |
| Sequencing by hybridization | Massachusetts General Hospital, Boston, Massachusetts/Unidad de Diagnóstico y Tratamiento de Enfermedades Metabólicas Congénitas, Hospital Clínico Universitario de Santiago/Harvard University |
| Sequencing with mass spectrometry | Columbia Genome Center, Columbia University College of Physicians and Surgeon |
| Microfluidic Sanger sequencing | Northwestern University/University of California |
| Microscopy-based techniques | Harvard University |
| RNAP sequencing | Institute of General and Molecular Biology, Nicolaus Copernicus University |
| In vitro virus high-throughput sequencing | The Institute of Medical Science, The University of Tokyo |
Table 2: Technologies under development in line of advanced NGS.
Massive parallel sequencing facility may help in understanding complex, elusive neurological and mental diseases such as schizophrenia, depression ,bipolar disorder (BD), obsessive compulsive disorder (OCD), post-traumatic stress disorder (PTSD), attention deficit, hyperactivity disorder (ADHD) etc.[16].
Diseases with multiple gene involvement and alteration in the condition due to the environmental and other relevant factors are still clinically inconclusive. NGS based studies with large number of samples with acceptable range of variations may help in finalizing the actual causative factors of such elusive mental disorders [17]. NGS based whole genome sequencing for complex human trait analysis with the help of larger cohort will support in overcoming the problems with reproducibility obtained through linkage and association based studies conducted for the psychiatric disorders [18].
Genome-wide Association Studies (GWAS)
Association of a particular disease with specified genes was earlier confirmed through linkage studies only [19]. This method was not much reproducible and at the same time, not able to associate multiple genes or genetic factor for a specific disease condition. Advanced sequencing technology, such as NGS and development of several essential genome based databases, especially with Single Nucleotide Polymorphism (SNP) based information with modern storage and retrieval system allowed to opt for Genome Wide Association Studies (GWAS) for different diseases [20-23]. The initial attempt in this direction in 2005 for the age related macular degeneration revealed the association of various genes which were not reported earlier along with pathophysiological insight related to complement-facilitated inflammation [24]. Later on, this revelation led towards having targeted therapeutic approach for this inflammation related pathway pertaining to macular degeneration [25].
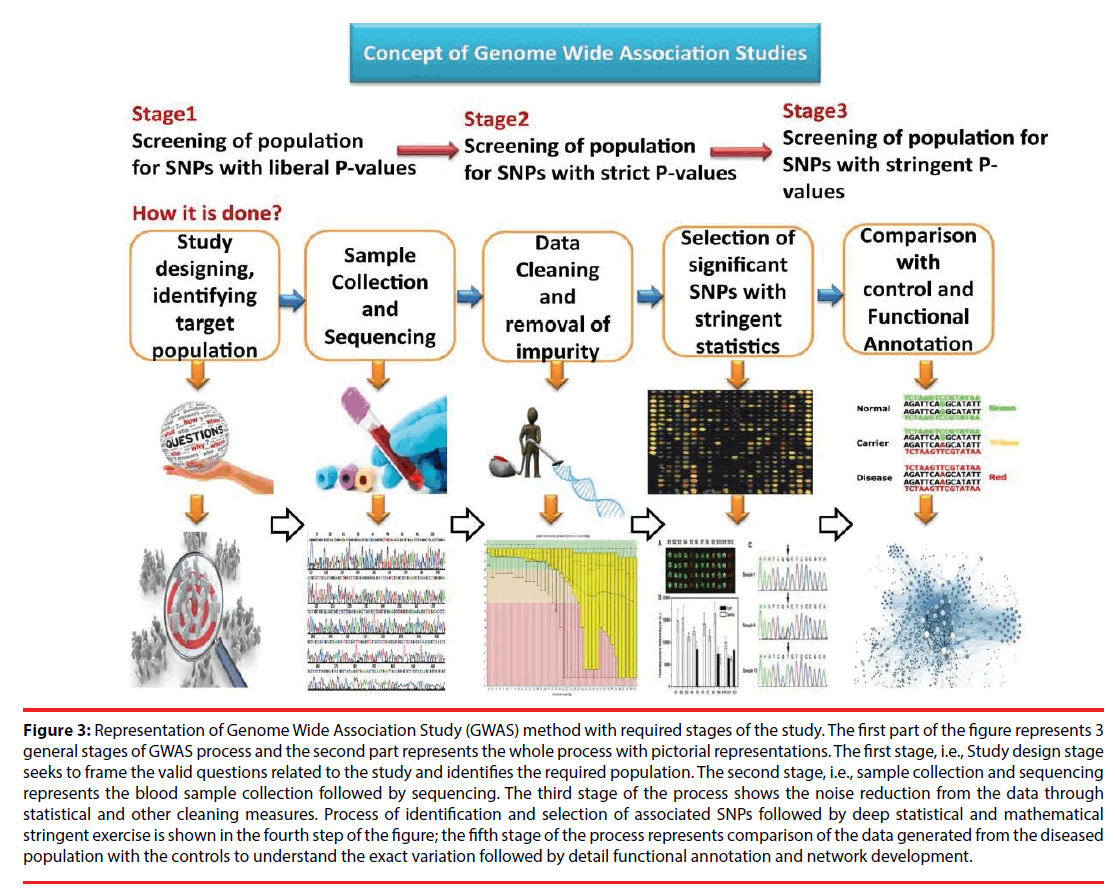
GWAS methodology considers sampling of blood samples, preferably collected from a large cohort [26]. In this phenotype-first approach, the complete sample is segregated into the diseased samples and the control. Sequencing of the samples is done through high throughput methods such as NGS and variation is determined among the diseased and the control genomes [27,28]. Thus, SNP frequency variation associated with the trait or disease under consideration is estimated followed by identification of the locations of such SNPs in the whole genome including coding, noncoding, and all other regions. Correlating each SNPs association with the disease trait remains onerous [29]. Moreover, it is not always possible to correlate each identified SNPs contribution towards occurrence of disease. Statistical measures such as Odd ratio and test of significance are used to gain information in this aspect. In the past few decades, this approach revealed enormous information pertaining to the search for genetic cause related to a specific disease. Similar technical approaches are adopted to understand the underlying genetic factors responsible for complex psychological and neurological disorders [30]. The general GWAS procedure is depicted in Figure 3.
Figure 3: Representation of Genome Wide Association Study (GWAS) method with required stages of the study. The first part of the figure represents 3 general stages of GWAS process and the second part represents the whole process with pictorial representations. The first stage, i.e., Study design stage seeks to frame the valid questions related to the study and identifies the required population. The second stage, i.e., sample collection and sequencing represents the blood sample collection followed by sequencing. The third stage of the process shows the noise reduction from the data through statistical and other cleaning measures. Process of identification and selection of associated SNPs followed by deep statistical and mathematical stringent exercise is shown in the fourth step of the figure; the fifth stage of the process represents comparison of the data generated from the diseased population with the controls to understand the exact variation followed by detail functional annotation and network development.
GWAS in Schizophrenia
Schizophrenia is a well-known complex mental disorder characterized by hallucinations, delusions, disordered thinking and behavior. The symptoms are positive, negative and cognitive types. Genetic issues including heritability are already known to be a factor for this condition along with changes in the brain structure and chemistry. Interaction between environment and genes is also known to be an important factor for triggering the onset and manifestation and progression of this disease condition.
As heritability and other genetic factors have been already associated with this disease, deep insight was obtained with several novel revelations through the genome wide associated studies. More than 20 genes have been found to be associated with this disease through this exercise and several genes were found repeatedly in many of the similar studies conducted so far [31]. The genes identified through GWAS methods also demonstrated considerable amount of statistical significance under stringent statistical evaluations [31]. Discovering the genes connected with the disease through GWAS analysis has become a continuous process but elucidation of functional association and its detailed mechanism is another complex part of such analysis which may provide valuable pathophysiological mechanism related information. As Schizophrenia is a polygenic disease, estimation of each gene’s contribution is not feasible; instead, exploring those genes which are involved and their mode of association with the disease will provide better enlightenment [32]. Efforts from different consortium such as International Schizophrenia Consortium (ISC), SGENE and Molecular Genetics of Schizophrenia (MGS) and Schizophrenia Psychiatric GWAS Consortium (PGC) received lot of attention where GWAS was implemented extensively [32-34]. Several interesting findings related to Schizophrenia were reported as a result of the large scale case control studies with large number of samples and haplotypes. Connection of MHC (Major Histocompatibility Complex) region with Schizophrenia was first reported in one of these studies [35]. Further analysis and meta-analysis revealed that neurogranin (NRGN, locus: 11q24.2) and transcription factor 4 (TCF4) genes were also associated with the disease condition apart from the MHC region [32-35]. Exploring further associations of other genomic regions also provided insight regarding the microRNA MIR137 which was found associated with Schizophrenia. GWAS studies also unveiled the association of TCF4 which plays role in nervous system development in many instances. Contribution of novel loci such as, for microRNA 137(MIR137), 1p23.3; Prostate-specific transcript 1(PCGEM1), 2q32.3;CUB and Sushi Multiple Domains 1(CSMD1), 8p23.2; Matrix Metallopeptidase 16 (MMP16), 8q21.3 and Cyclin and CBS Domain Divalent Metal Cation Transport Mediator 2/5’-Nucleotidase, Cytosolic II (CNNM2/NT5C2), 10q24.32-q24.33 in disease progression was discovered through these studies [36-40]. Population specific GWAS in relation to Schizophrenia has also been conducted recently. Analysis of Chinese Schizophrenic patients revealed relation of 11p11.2 locus, 8p12 and 1q24.2 loci with the disease [41].
Apart from association of loci, copy number variants (CNVs) are also linked with Schizophrenia. Particular longer stretches of nucleotides which were either deleted or duplicated are considered as copy number variants (CNVs) [42]. Such eight important variant regions present in the genome favoring Schizophrenia were reported by Mowry and Gratten [43]. Most of these variants showed greater Odd ratios and are found rarely in nature. Among these, CNVs deletions and duplications both were observed. Direct relation of CNVs with Schizophrenia is not yet clear but related studies reported that certain percentage of CNVs might be associated with either Schizophrenia onset or progression [44].
GWAS in Epilepsy
Epilepsy is defined as a neurological condition with symptoms including epileptic seizures which are tonic-clonic seizures sometimes often having absence of seizures also. Even though there is a large array of Anti-epileptic drugs (AEDs) available to control or reduce the condition yet the proper genetic cause is not known [45]. Epilepsy is heritable in nature and genetic aspects are associated with this neurological condition [45]. Interestingly, Epilepsy is a polygenic disease which involves multiple genes. The success rate of GWAS based studies in identification of risk associated loci pertaining to Epilepsy is low due to the small sample size as well as effect size for polygenic disease. Cohort studies with patients having epilepsy with remission of seizure due to followed treatment regime were carried out recently [46]. Investigation of the SNPs involved along with the identification of the statistically evaluated involvement of loci suggested that 6p12.2, 9p23 and 15q13.2 are associated with epilepsy which are related to PTPRD (Receptor-type tyrosineprotein phosphatase delta), ARHGAP11B (Rho GTPase Activating Protein 11B), having key role in amplification of basal progenitor and vital for the expansion of neocortex) and GSTA4 (Glutathione S-Transferase Alpha 4) which is responsible for Atherosclerosis. Analysis of pathways suggested that ‘calcium signaling’ and ‘phosphatidylinositol signaling pathways are associated with epileptic conditions [46]. Reproducibility remained one of the major hurdles in GWAS based analysis of epileptic data. Recently, incorporation of advanced statistical measures and large number of samples have been reported in this context [47]. Lack of large number of cases with specific predominant neurological conditions inspired researchers to seek solutions from different species or other animal models. It is stated that ADAM23 (a disintegrin and metalloprotease domain 23) gene which interacts with LGI1 (Leucine-rich, glioma inactivated 1) and LGI2 (Leucine-rich, glioma inactivated 2) protein is related to epilepsy [48]. Twin studies and population based studies provided useful insight towards the epilepsy. During assessment of genetic syndromes of epilepsy, monozygous twins were found to be more consistent than dizygous [49]. Large twin studies in Danish population for epileptic conditions suggested that genetic factors contributes substantially for the disease etiology, Further heritability of the disease condition was found high and incremental with age [50].
GWAS in Parkinson’s Disease
Similar to the other neurological disorders, Parkinson disease has also emerged as a major health and social issue in the recent times. There are overlapping symptoms with other similar neurological disorders which include whole body issues, muscular and tremor associated problems, disturbed sleep, impaired speech, anxiety, lack of smell detection, urinary issues, and uncontrolled facial expression etc. Reports suggest that 1-2 individuals out of 1000 are vulnerable to the disease condition [51]. This disease claims at least 1% of the population having age over 60 years [51]. Thus, detailed investigation of the disease mechanism and relation with age is crucial. GWAS based studies have been successful in unraveling information pertaining to PD. Relation and significance of PD causing genes, such as α-synuclein and LRRK2 (Leucine-rich repeat kinase 2) have been established through GWAS [52]. Stratified GWAS analysis further identified novel loci in the short arms of chromosome 1 and 8, which are probably associated with PD [53]. Different loci have been identified through GWAS protocol in various population groups such as Ashkenazi Jewish population [54] and Dutch population [55], thus helping in enhancing our present understanding with relation to different population groups and identifying population specific Parkinson associated markers. During this process of exploration through analysis and meta-analysis, multiple important loci were found to be actively associated with the PD such as RIT2 (Ras Like Without CAAX 2) [56], GBA (Glucosylceramidase Beta), SNCA (Synuclein Alpha), HLA (Major Histocompatibility Complex), GAK/DGKQ (Cyclin G Associated Kinase/Diacylglycerol Kinase Theta) [57,58], LRRK2 (Leucine-rich repeat kinase 2), MAPT (Microtubule Associated Protein Tau), SCARB2 (Scavenger Receptor Class B Member 2), SREBF1/RAI1 (Sterol Regulatory Element Binding Transcription Factor 1/Retinoic Acid Induced 1) [59], PARK16(Parkinson Disease 16 (Susceptibility))/1q32, STX1B (Syntaxin 1B)/16p11, FGF20 (Fibroblast Growth Factor 20)/8p22, STBD1 (Starch Binding Domain 1)/4q21, and GPNMB(Glycoprotein Nmb)/7p15 [60,61]. Identification of all the associated loci for this neurological condition is important to have a deep understanding for this disease as well as population specific genomic condition.
GWAS in Alzheimer’s Disease
Alzheimer’s disease is majorly reported in the elderly population above 60 years of age with the classical symptom of memory loss. At present, globally the disease is reported to affect other age groups also. Initially, the disease was mistaken as part of the natural aging process, but now it is an established fact that losing memory up to the level where the regular tasks are hindered is a disease condition. Clinically, the condition progresses through early or mild, moderate and severe stage. Treatment depends on the stage of disease. Even though symptomatic treatment is available yet there is no specific cure available at present.
Several studies identified the genes responsible for Alzheimer. It was established that genes such as Amyloid-beta A4 protein precursor (APP), presenilin 1 (PSEN1), and presenilin 2 (PSEN2) are directly associated with Alzheimer disease. The complex relation between these proteins in relation to Alzheimer is interesting where Amyloid Precursor Protein (APP) is directly involved with Alzheimer and PSEN proteins are related to the processing of γ-secretase which further plays important role in processing the APP protein. APP is involved in synapse formation, maintenance of neural plasticity and exporting of iron molecules. All these proteins are mainly involved in up-regulation and downregulation of several membrane proteins. Genetic inheritance of Alzheimer has been established earlier where apart from the strong evidence of inheritance; higher probability in the specific offspring of the carriers is also discussed [62,63]. Generally, Alzheimer is clinically categorized as early-onset Alzheimer and late-onset Alzheimer disease. Mutations in the previously mentioned genes are commonly observed in the early onset cases of Alzheimer whereas presence of APOE ε4 alleles with greater frequency has been reported in case of late onset of Alzheimer [64,65]. Multiple linkage studies with reference to Alzheimer have been carried out majorly depending on the microsatellite based markers [66]. These linkage studies established the role of Apolipoprotein E (ApoE) or APOE region which is present in the chromosome 9 for late onset Alzheimer diseases (LOAD). Other specific genomic regions were also identified through the marker based linkage studies which were found strongly associated with Alzheimer disease.
GWAS based analysis have been extremely helpful in establishing this disease condition and investigating the genome based cause and identifying the involved genomic regions [67]. SNP dependent GWAS analyses revealed several other important genetic factors related to Alzheimer disease in the later stage of scientific investigations. Samples were collected and utilized to understand LOAD from US, UK, Canada, Germany, Netherlands and other geographical regions and studies were conducted on a case control or family basis. Numerous specific genes were found to be associated or with plausible association with the disease condition during these extensive investigations. Some of those genes and specific chromosomal regions identified with higher statistical significance were Galanin Like Peptide (GALP), Lamin A/C (LMNA), PiggyBac Transposable Element Derived 1 (PGBD1), Thioesterase Superfamily Member 5 (THEM5), Myosin Heavy Chain 13 (MYH13), Trafficking Kinesin Protein 2 (TRAK2),Early B-Cell Factor 3 (EBF3) [68], GRB2 Associated Binding Protein 2 (GAB2) [69], Golgi Phosphoprotein 2 (GOLPH2), chr9p24.3, chr15q21.2 [70], Lecithin Retinol Acyltransferase (LRAT) [71], Microtubule Associated Protein Tau (MAPT), Sortilin Related Receptor 1 (SORL1), Cholinergic Receptor Nicotinic Beta 2 Subunit (CHRNB2), Cholesterol 25-Hydroxylase (CH25H), Phosphoenolpyruvate Carboxykinase 1 ( PCK1) [72]. Apart from these, several other genes were found to be associated with LOAD but not much information on their roles is available so far. Reports on specific studies have been summarized by Bertram and Tanzi [73], Tosto and Reitz [74] and recently by Shen and Jia [75].
GWAS in Bipolar Disorder
Manic depression or bipolar disorder is a growing psychological and behavioral problem in the stress filled competitive life. The common symptoms are requirement of less sleep, ignoring real time situations, loss of interest in regular activities, hyper or lower energy depending on the episodes patient is going through at the moment. So far, causative factors have not been established for this condition. It is speculated that multiple factors including genetic make-up, altered brain structure, altered physiochemical balance of the regular elements and fluids in the brain are responsible for this disorder in general. Other than infants and kids, this disease affects all the age groups. Middle and elder age group are the most vulnerable age group for this condition. Estimates suggest that almost 1% of the global population suffers from bipolar disorder [76]. Surprisingly, 3% of the population faces this condition in one or the other phase of their life [77]. The overall episodes are segregated into manic, hypomanic and depressive episodes. The initial genetic linkage of this disease condition was established in the late 60s [78] through study of the family histories. Other investigations revealed several polymorphisms associated with genes such as TPH1 (Tryptophan Hydroxylase 1), BDNF (Brain Derived Neurotrophic Factor), DAO (D-Amino Acid Oxidase), DRD4 (Dopamine Receptor D4) etc. have been also reported [79]. To further understand the genetics associated with the disease, large cohort based GWAS studies involving 40,000 individuals have been conducted [80]. The study revealed association of two specific loci with the disease including intergenic region at position of 9p21.3 and ERBB2 gene associated marker [80]. But the study failed to connect the association of the X chromosome or other strong evidence related to the bipolar condition. Other such risk loci associated with the bipolar disorder found were specific markers near to Diacylglycerol Kinase Eta (DGKH) [81], CACNA1C (Calcium Voltage-Gated Channel Subunit Alpha1 C) [82], TENM4 (Teneurin Transmembrane Protein 4), ADCY2 (Adenylate Cyclase 2), SYNE1 (Spectrin Repeat Containing Nuclear Envelope Protein 1), ANK3 (Ankyrin 3), TRANK1(Tetratricopeptide Repeat And Ankyrin Repeat Containing 1) genes [83]. Gene cluster found in the chromosome 3p21was also reported to have a strong link-up with the disease condition [84-87]. Scientific evidence revealed the relation between the altered brain function due to change in the regional brain activity and BD. Further investigation suggested that polygenic risk factors associated with the bipolar disorder impacts the visual ability and processing of normal emotion of the patients. This information associated with bipolar disorder was acquired from two different studies employing different scientific approaches, through GWAS [88] and neuro imaging [89]. Apart from association of BD with several genes such as SORCS2 (Sortilin Related VPS10 Domain Containing Receptor 2), DFNBP31 (Deafness Type 31 Protein) and SLC39A3 (Solute Carrier Family 39 Member 3), it was observed [88] that bipolar type 1 disorder is related with the SNPs found close to CDH7 (Cadherin 7) gene. Interestingly, this CDH7 gene is part of the cadherin superfamily which aids in development of the brain in multiple ways including axon growth, cell sorting and other relevant roles. Cross population genome investigations in relation to the bipolar disorder in European and Japanese population also provided important clue that Japanese population shows presence of susceptible locus at chromosomal position 11q12.2, this region is known to have regulatory genes viz.FADS1 (Fatty Acid Desaturase 1), FADS2(Fatty Acid Desaturase 2) and FADS3((Fatty Acid Desaturase 3). Another gene found associated with BD was NFIX (Nuclear Factor IX) [90]. Another study carried out by Mühleisen et al. [87] including Psychiatric GWAS Consortium Bipolar Disorder Working Group suggested the possible link of loci in vicinity to ODZ4 (Odd Oz/Ten-M Homolog 4 or Teneurin Transmembrane Protein 4) gene with bipolar disorder [84]. Connection between tissue content depletion in bipolar disorder and possible response of the patients to only effective therapy based on lithium was found through glutamatergic network mediation through in silico pharmacogenomics approach [91]. Further experimental insight is still required to confirm such molecular relation.
GWAS in Attention Deficit Hyperactivity Disorder (ADHD)
Along with other neurological conditions, there has been an increase in cases of Attention Deficit Hyperactivity Disorder (ADHD). This is another mental condition which was ignored and not even considered as a disorder till a very long time. This condition is chronic in nature and includes lack of detail attention, impulsive behavior in a stressed condition as well as hyperactivity. The population found to have this kind of disorder is majorly children population in pubertal stage and the middle aged people. But, this condition was also recorded in other age groups as well. Apparently, from clinical point of view, this medical condition might not have gained enough attention but the social impact of the condition is enormous which include accidents, injuries, suicide, criminal like behaviors, substance abuse etc. Presently, with growing number of cases worldwide, ADHD is receiving serious attention from the medical practitioners as well as researchers considering the far reaching social impact of the disorder. We have very limited knowledge about this condition and a proper cure is still evading us. It was observed that parents who demonstrate few symptoms of ADHD are more likely to pass on the condition to their children [92,93]. Therefore, genetic link was established in the early 2000 for ADHD. Considering the genetic basis, multiple studies have been conducted to search out the responsible and associated genes. Results from various studies suggest that catecholaminergic genes such as dopamine beta-hydroxylase (DBH), dopamine D4 receptor (DRD4), dopamine D5 receptor (DRD5), synaptosomal-associated protein (SNAP-25), serotonin transporter(SLC6A4) and 5-Hydroxytryptamine Receptor 1B (HTR1B) genes are associated with ADHD.
Genome wide association studies helped to understand the related gene network and their functional influence on the condition. Genetic correlations were found with several phenotypes [94] such as depressive disorder, mortality, educational outcomes etc. Investigations suggest Forkhead box protein P2 (FOXP2) gene region which is important for the development of speech and language and Dual Specificity Phosphatase 6(DUSP6) gene associated loci may be responsible for the etiology of the ADHD [94]. Recently, Stergiakouli and coworkers confirmed the relation between polygenic score of ADHD and educational outcomes and cognition for common population [95]. Combined GWAS and protein-protein analysis of Obsessive Compulsive Disorder (OCD) and ADHD revealed that none of the SNPs investigated were above the significance cutoff, but SNP rs10989904 was found to have the strongest possible association [96]. From various studies, several other genes were also found to be associated with ADHD such as CDH13 (Cadherin 13), ASTN2 (Astrotactin 2), CTNNA2 (Catenin Alpha 2), KALRN (Kalirin RhoGEF Kinase), PRKG1 (Protein Kinase, CGMP-Dependent, Type I), FLNC (Filamin C), TCERG1L (Transcription Elongation Regulator 1 Like), PPM1H (Protein Phosphatase, Mg2+/ Mn2+ Dependent 1H), NXPH1 (Neurexophilin 1), PPM1H (Protein Phosphatase, Mg2+/ Mn2+ Dependent 1H), HK1 (Hexokinase 1), HKDC1 (Hexokinase Domain Containing 1), GRM5 (Glutamate Metabotropic Receptor 5), CHRNA7 (Cholinergic Receptor Nicotinic Alpha 7 Subunit), XKR4 (XK Related 4), FAM190A (Family With Sequence Similarity 190, Member A or Coiled-Coil Serine Rich Protein 1), SLC9A9 (Solute Carrier Family 9 Member A9), CHMP7 (Charged Multivesicular Body Protein 7), TNFRSF10D (TNF Receptor Superfamily Member 10d), TNFRSF10A (TNF Receptor Superfamily Member 10a) and LOXL2 (Lysyl Oxidase Like 2) [97]. Even though several GWAS case-control studies were conducted to understand the genetic causes of ADHD along with population specific studies [98], they are not conclusive for confirming multiple genetic factors responsible for the condition.
GWAS in Amyotrophic Lateral Sclerosis (ALS)
ALS or Amyotrophic Lateral Sclerosisis is also known as Lou Gehrig’s disease or motor neuron disease (MND). The medical condition manifests as progressive muscle weakness in the patient and the disease is rare. With time, neurons controlling the voluntary muscles become inactive and unresponsive towards any stimulation. Decrease in muscle mass, increase in muscle stiffness and twitching are some of the common symptoms for this condition. In most of the cases, the actual cause of the disease onset or progression is unknown. ALS is broadly classified into sporadic ALS (SALS) and familial ALS (FALS) based on the family history available for the considered cases. The former form of ALS accounts to almost 90% of the total cases whereas the latter affects only 10% of the cases. Genetic links pertaining to ALS have been discovered where autosomal dominant transmission is commonly observed along with onset in adults. On the contrary, recessive dominance is observed with juvenile onset of the disease conditions with specific mutation in superoxide dismutase (SOD1) gene [99]. Besides, other genes found associated with adult FALS were TAR DNA-binding protein (TDP-43), Ataxin 2 (Atx2), fused in sarcoma/ translocated in liposarcoma (FUS/TLS), vesicleassociated membrane protein associated protein B (VAPB), angiogenin (ANG) and valosincontaining protein (VCP) [100]. Similarly, genes associated with the juvenile onset of ALS are Alsin, Senataxin (SETX) and Spatacsin (SPG11). Several other genes were also associated with the rare form of the disease such as Ubiquilin-2 (UBQLN2). Some genes were suspected to have an implication in ALS onset including profilin-1 (PFN1), FIG4, matrin-3 (MATR3), Sigma Non-Opioid Intracellular Receptor (SIGMAR), v-erb-b2, erythroblastic leukemia viral oncogene homolog 4 and (ERBB4) genes [100]. Apart from MND, frontotemporal dementia (FTD) was also found to be strongly associated with ALS [101].
Genome-wide Complex Trait Analysis (GCTA)
Various consortium based collective efforts are being adopted for psychiatric disorder associated gene discovery, functional annotation and assessment of the genomic regions involved in psychiatry related issues and precision mapping of the important loci etc.[16].
Active sample analyses are being carried out for Schizophrenia, Autism, Bipolar disorder, MDD (Major Depressive Disorder), ADHD (Attention Deficit Hyperactive Disorder), Anorexia nervosa, OCD, Alzheimer’s disease and PTSD (Post- Traumatic Stress Disorder) etc.
Rigorous investigations related to gain-offunction and loss-of-function related to neurodevelopmental disorders have been carried out. Significant impact of mono-allelic expression (MAE) in comparison to bi-allelic expression (BAE) genes in this relevance has been reported [17].
Human well-being is one of the most complex topics under the realm of psychology and multiple hypothesis and complex criteria have been proposed to describe and quantify the same. Numerous analyses, evaluation and reevaluation are going on in this direction; majorly focusing on genome data based analysis. Identification and quantification of the selected genes which may have impact on well-being as per the Hedonic or Eudaimonic view have gained momentum in the recent times. Various effective hypotheses have been formulated to correlate the relations from a functional genomics perspective [102-104].
Medical Ethics for Psychiatric Disease Analysis through Genomics
Practicing science always had the goal of benefitting the human society and intended towards developing an excellent, cultured, civilization respecting and protecting the purpose of all life forms. Ethical consideration has always remained prime concern during all research activities. With time, concerns pertaining to the ethical issues have been raised and the existing protocols have been modified from time to time depending on the continued learning and inputs from the experts. Moreover, amendments in the presently followed regulations and guidelines become mandatory considering the scientific advancements and practices. Strict regulations have been framed for GMOs (genetically modified organisms), public health related research, pesticides, herbicides, clinical research and other aspects. As human, we must be concerned or should at least follow some ethical barrier depending on the situation.
Since last decade, genetic testing and genome based analysis have raised the major concerns due to the potential scientific and business practices and plausible misinterpretation and misuse of the obtained results. Several ethical questions are being asked by the scientific and social communities and strict rules are demanded by the concerned authorities.
In connection to clinical and genome based research, properly obtained informed consent remained a world-wide issue [105]. Similarly, data generation and data sharing with permission is another important issue [106]. Disease outbreak, especially viral and other infectious diseases also require proper ethical concerns and protocol in relation to quarantine, testing or application of unapproved medication during any emergency situation [107].
Need of strong ethical guidelines is often observed in the biomedical context considering the research aspects as well as the business perspectives. Primary care and genetic analysis have come closer with time. This is the time when we should consider patient’s safety with great caution and regard as much as possible [108].
Studies and research pertaining to neuroscience have always remained under the scanner of ethical committee guidelines while dealing with human or animal subjects. Considering the critical phases of treatments that the patients or subjects undergo, guidelines have become strict. Often, the marked border lines become blurred due to one or the other decision making process by the investigators. Thus, it becomes tough to justify the situation and the decision taken time and again.
Moreover, involvement of genomics related aspects compelled the scientific community and medical practitioners to raise concern. More rational and careful approach that ensures selection of the participants, collection of adequate number of samples, repetition of the experiments for the required number of times for a conclusive result, unbiased participants irrespective of the socio-economic background and presentation of impartial outcomes is the need of the time [109]. In case of neurological disorders, where it is often cumbersome to distinguish between a normal healthy and diseased condition, ethical issues play a major role. Moreover, critical surgeries associated with brain also require proper consent from the patient party as well as from the ethics regulation committee [110].
Considering the latest development of NGS, GWAS based technologies; several questions need to be addressed. Especially, for NGS technique implementation towards diagnostics applications, reporting protocol, incidental discoveries and variants of uncertain significance (VUS) lack clarity [111]. Arguments have been posed on the access of the data by the study participants themselves [112] and the fate of the unsolicited findings [113]. Another important issue is the difference in regulatory rules across various geographical regions. Rules vary from country to country and so far, global harmonization is not witnessed in several aspects. Country specific as well as global uniform rules and regulations pertaining to the NGS data analysis and interpretations are warranted [114].
Conclusion and Future Perspective
Neurological diseases are considered intriguing and complex and understanding these diseases remains a priority in medical science. Numerous attempts were made to gain some valuable insight using computational approaches for Parkinson’s disease and ALS [115,116]. Further we assembled and reviewed information on neurochemicals and their impact on behavior [117]. Gaining knowledge about the neural system can help in defining who we are and how we are functioning. Several intriguing and unanswered questions continue to confound research community. Technical advances in the recent times in this direction provided us the impetus to move a step ahead and investigate those dark corners which remained unexplored till date. Complexity of neurological and psychological conditions is one of the most critical enigmas of medical sciences. Distinguishing between normal and disease condition for certain diseases has been a great challenge to the medical sciences, leave alone exploring the in-depth causes. The latest techniques including Next Generation Sequencing (NGS), Genome-Wide Association Studies (GWAS), and Genome-wide Complex Trait Analysis (GCTA) have revolutionized the way we approach these problems and are expected to extend our present knowledge base regarding the general brain function and its deviation during neurological or psychiatric disease conditions. It is expected that in near future we will have a better understanding and safe and secure established treatment protocol for critical disease conditions such as Alzheimer, Parkinson, Epilepsy, Bipolar, ALS, and ADHD.
The pattern of scientific practice has to changed with time depending on the need. At times, scientific research was esoteric in nature. Only concerned members from the related fraternity were allowed o participate in certain research rograms and have access to the relevant information and research outcomes. Presently, many communities are gradually getting involved in research programs, either as subjects or contributors. Improvement in mode of communication and access to research amenities helped all to participate in mass research programs. We should define limitation of access depending on the far reaching impact of research and development. A balance between the ethical conditions and technical implementation has become a sine qua non. Therefore, a collective effort from the researchers, participants and funding agencies may facilitate in developing a better and safe system for unfolding the mysteries of the nature and provide effective solutions for the neurological disease conditions.
Acknowledgement
Neelima Arora thanks University Grants Commission for PostDoctoral Fellowship
References
- Plomin R, Colledge E. Genetics and psychology: beyond heritability. Eur. Psychol 6(4), 229-240 (2001).
- World Health Organization. Neurological disorders: public health challenges. World Health Organization (2006).
- Sachdev PS, Blacker D, Blazer DG, et al. Classifying neurocognitive disorders: the DSM-5 approach. Nat. Rev. Neurol 10(11), 634-642 (2014).
- Plomin R, Deary IJ. Genetics and intelligence differences: Five special findings. Mol. Psychiatry 20 (1), 98-108 (2015).
- Spearman C. The abilities of man: McMillan and Co.Ltd. London (1927).
- Selzam S, Krapohl E, Von Stumm S, et al. Predicting educational achievement from DNA. Mol. Psychiatry 22(2), 267-272 (2017).
- Bachman DL, Green RC, Benke KS, et al. Comparison of Alzheimer’s disease risk factors in white and African American families. Neurology 60(1), 1372-1374 (2003).
- Pastor P, Ezquerra M, Tolosa E, et al. Further extension of the H1 haplotype associated with progressive supranuclear palsy. Mov. Disord 17(1), 550-556 (2002).
- Jackson JF, Currier RD, Teresaki PI, et al. Spinocerebellar ataxia and HLA linkage: risk prediction by HLA typing. N. Engl. J. Med 296(1), 1138-1141 (1997).
- Bird TD, Ott J, Giblett ER. Evidence for linkage of Charcot-Marie-Tooth neuropathy to Duffy locus on chromosome 1. Am. J. Hum. Genet 34(3), 388-394 (1982).
- Pulst SM. Neurogenetics: single gene disorders. J. Neurol. Neurosurg. Psychiatry 74(12), 1608-1614 (2003).
- Yang J, Lee SH, Goddard ME, et al. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet 88(1), 76-82 (2011).
- Green ED, Watson JD, Collins FS. Human Genome Project: Twenty-five years of big biology. Nature 526(7571), 29-31 (2015).
- Quail MA, Smith M, Coupland P, et al. A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC. Genomics 13(1), 341 (2012).
- Liu L, Li Y, Li S, et al. Comparison of next-generation sequencing systems. J. Biomed. Biotechnol 251364 (2012).
- Senthil G, Dutka T, Bingaman L, et al. Genomic resources for the study of neuropsychiatric disorders. Mol. Psychiatry 22(12), 1659-1663 (2017).
- Savova V, Vinogradova S, Pruss D, et al. Risk alleles of genes with monoallelic expression are enriched in gain-of-function variants and depleted in loss-of-function variants for neurodevelopmental disorders. Mol. Psychiatry 22(12), 1785 (2017).
- Churchill GA, Airey DC, Allayee H, et al. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nature. Genetics 36(11), 1133-137 (2004).
- Wu Z, Wang N, Murong S, et al. Carrier detection and presymptomatic identification of Wilson disease in Chinese by non-isotopic linkage analysis with four short tandem repeat polymorphisms. J. Tongji. Med. Univ 19(1), 50-65 (1999).
- Yonan AL, Alarcón M, Cheng R, et al. A Genomewide Screen of 345 Families for Autism-Susceptibility Loci. Am. J. Hum. Genet 73(4), 886-897 (2003).
- de Moor MHM, van den Berg SM, Verweij KJH, et al. Meta-analysis of Genome-wide Association Studies for Neuroticism, and the Polygenic Association With Major Depressive Disorder. JAMA. Psychiatry 72(7), 642 (2015).
- Verweij KJH, Vinkhuyzen AAE, Benyamin B, et al. The genetic aetiology of cannabis use initiation: A meta-analysis of genome-wide association studies and a SNP-based heritability estimation. Addict. Biol 18(5), 846-850 (2013).
- Devanna P, Chen XS, Ho J, et al. Next-gen sequencing identifies non-coding variation disrupting miRNA-binding sites in neurological disorders. Mol. Psychiatry 23(5), 1375-1384 (2017).
- Klein RJ, Zeiss C, Chew EY, et al. Complement factor H polymorphism in age-related macular degeneration. Science 308(5720), 385-389 (2005).
- Rohrer B, Long Q, Coughlin B, et al. A targeted inhibitor of the alternative complement pathway reduces angiogenesis in a mouse model of age-related macular degeneration. Invest. Ophthalmol. Vis. Sci 50(7), 3056-3064 (2009).
- Craig JE, Hewitt AW, McMellon AE, et al. Rapid inexpensive genome-wide association using pooled whole blood. Genome. Res 19(11), 2075-2080 (2009).
- Lowe WL, Reddy TE. Genomic approaches for understanding the genetics of complex disease. Genome. Res 25(10), 1432-1441(2015).
- Kilpinen H, Barrett JC. How next-generation sequencing is transforming complex disease genetics. Trends. Genet 29(1), 23-30 (2013).
- Turner S, Armstrong LL, Bradford Y, et al. Quality Control Procedures for Genome Wide Association Studies. Curr. Protoc. Hum. Genet Chapter 1(Unit1), 19 (2011).
- Bras J, Guerreiro R, Hardy J. Use of next-generation sequencing and other whole-genome strategies to dissect neurological disease. Nat. Rev. Neurosci 13(7), 453-464 (2012).
- Bergen SE, Petryshen TL. Genome-wide association studies of schizophrenia: Does bigger lead to better results? Curr. Opin. Psychiatry 25(2), 76-82 (2012).
- Purcell SM, Wray NR, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 460(7256), 748-752 (2009).
- Sullivan PF. The psychiatric GWAS consortium: big science comes to psychiatry. Neuron 68(2), 182-186 (2010).
- Logue MW, Amstadter AB, Baker DG, et al. The Psychiatric Genomics Consortium Posttraumatic Stress Disorder Workgroup: Posttraumatic Stress Disorder Enters the Age of Large-Scale Genomic Collaboration. Neuropsychopharmacology 40(10), 2287-2297 (2015).
- Mokhtari R, Lachman HM. The Major Histocompatibility Complex (MHC) in Schizophrenia: A Review. J. Clin. Cell. Immunol 7(6), 479 (2016).
- Harrison PJ. Recent genetic findings in schizophrenia and their therapeutic relevance. J. Psychopharmacol 29(2), 85-96 (2015).
- Shi J, Levinson DF, Duan J, et al. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature 460(7256), 753-757 (2009).
- Stefansson H, Ophoff RA, Steinberg S, et al. Common variants conferring risk of schizophrenia. Nature 460(7256), 744-747 (2011).
- Ripke S, Sanders A, Kendler K. Genome-wide association study identifies five new schizophrenia loci. Nature 43(10), 969-976 (2011).
- Shi Y, Li Z, Xu Q, et al. Common variants on 8p12 and 1q24.2 confer risk of schizophrenia. Nat. Genetics 43(12), 1224–1227 (2011).
- Yue WH, Wang HF, Sun LD, et al. Genome-wide association study identifies a susceptibility locus for schizophrenia in Han Chinese at 11p11.2. Nat. Genet 43(12), 1228-1231 (2011).
- Kirov G, Rees E, Walters JTR, et al. The penetrance of copy number variations for schizophrenia and developmental delay. Biol. Psychiatry 75(5), 378-385 (2014).
- Mowry BJ, Gratten J. The emerging spectrum of allelic variation in schizophrenia: Current evidence and strategies for the identification and functional characterization of common and rare variants. Mol. Psychiatry 18(1), 38-52 (2013).
- Costain G, Lionel AC, Merico D, et al. Pathogenic rare copy number variants in community-based schizophrenia suggests a potential role for clinical microarrays. Hum. Mol. Genet 22(22), 4485-4501 (2013).
- Speed D, O’Brien TJ, Palotie A, et al. Describing the genetic architecture of epilepsy through heritability analysis. Brain 137(10), 2680-2689 (2014).
- Speed D, Hoggart C, Petrovski S, et al. A genome-wide association study and biological pathway analysis of epilepsy prognosis in a prospective cohort of newly treated epilepsy. Hum. Mol. Genet 23(1), 247-258 (2014).
- Poduri A. Meta-analysis revives genome-wide association studies in epilepsy. Epilepsy. Curr 15(3), 122-123 (2015).
- Seppälä EH, Koskinen LLE, Gulløv CH, et al. Identification of a novel idiopathic epilepsy locus in Belgian Shepherd dogs. PLoSONE 7(3) (2012).
- Berkovic SF, Howell RA, Hay DA, et al. Epilepsies in twins: Genetics of the major epilepsy syndromes. Ann. Neurol 43(4), 435-445 (1998).
- Kjeldsen MJ, Kyvik KO, Christensen K, et al. Genetic and environmental factors in epilepsy: a population-based study of 11900 Danish twin pairs. Epilepsy. Res 44(2-3), 167-178 (2001).
- Tysnes OB, Storstein A. Epidemiology of Parkinson’s disease. J. Neural. Transm 124(8), 901-905 (2017).
- Chai C, Lim K-L. Genetic insights into sporadic Parkinson’s disease pathogenesis. Curr. Genomics 14(8), 486-501 (2013).
- Hill-Burns EM, Wissemann WT, Hamza TH, et al. Identification of a novel Parkinson’s disease locus via stratified genome-wide association study. BMC. Genomics 15(1), (2014).
- Liu X, Cheng R, Verbitsky M, et al. Genome-Wide association study identifies candidate genes for Parkinson’s disease in an Ashkenazi Jewish population. BMC. Medical. Genetics 12(1), (2011).
- Simón-Sánchez J, Van Hilten JJ, Van De Warrenburg B, et al. Genome-wide association study confirms extant PD risk loci among the Dutch. Euro. J. Human Genetics 19(6), 655 (2011).
- Pankratz N, Beecham GW, Destefano AL, et al. Meta-analysis of Parkinson’s disease: Identification of a novel locus, RIT2. Ann. Neurol 71(3), 370-384 (2012).
- Nalls MA, Pankratz N, Lill CM, et al. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat. Genetics 46(9), 989-993 (2014).
- Edwards TL, Scott WK, Almonte C, et al. Genome-Wide association study confirms SNPs in SNCA and the MAPT region as common risk factors for parkinson disease. Ann. Human. Genetics 74(2), 97-109 (2010).
- Do CB, Tung JY, Dorfman E, et al. Web-based genome-wide association study identifies two novel loci and a substantial genetic component for Parkinson’s disease. PLoS. Genetics 7(6), (2011).
- Keller MF, Saad M, Bras J, et al. International Parkinson’s Disease Genomics Consortium (IPDGC) Wellcome Trust Case Control Consortium 2 (WTCCC2): Using genome-wide complex trait analysis to quantify ‘missing heritability’in Parkinson’s disease. Hum. Mol. Genet 21(4996), e5009 (2012).
- A Two-Stage Meta-Analysis identifies several new loci for Parkinson’s disease. PLoS. Genetics 7(6), (2011).
- Lautenschlager NT, Cupples LA, Rao VS, et al. Risk of dementia among relatives of Alzheimer's disease patients in the MIRAGE study What is in store for the oldest old?. Neurology 46(3), 641-650 (1996).
- Gatz M, Reynolds CA, Fratiglioni L, et al. Role of genes and environments for explaining Alzheimer disease. Arch. Gen. Psychiatry 63(2), 168-174 (2006).
- Corder E, Saunders A, Strittmatter W, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science 261(5123), 921-923 (1993).
- Bertram L, McQueen MB, Mullin K, et al. Systematic meta-analyses of Alzheimer disease genetic association studies: The AlzGene database. Nat. Genetics 39(1), 17-23 (2007).
- Ertekin-Taner N. Genetics of Alzheimer’s disease: A Centennial Review. Neurologic. Clinics 25(3), 611–667 (2007).
- Ertekin-Taner N. Genetics of Alzheimer disease in the pre- and post-GWAS era. Alzheimer’s. Res. Ther 2(1), (2010).
- Grupe A, Abraham R, Li Y, et al. Evidence for novel susceptibility genes for late-onset Alzheimer’s disease from a genome-wide association study of putative functional variants. Human. Mol. Genetics 16(8), 865-873 (2007).
- Reiman EM, Webster JA, Myers AJ, et al. GAB2 Alleles Modify Alzheimer’s Risk in APOE ε4 Carriers. Neuron 54(5), 713-720 (2007).
- Li H, Wetten S, Li L, et al. Candidate single-nucleotide polymorphisms from a genomewide association study of Alzheimer disease. Arch. Neurol 65(1), 45-53 (2008).
- Abraham R, Moskvina V, Sims R, et al. A genome-wide association study for late-onset Alzheimer’s disease using DNA pooling. BMC. Med. Genomics 1(1), 44 (2008).
- Feulner TM, Laws SM, Friedrich P, et al. Examination of the current top candidate genes for AD in a genome-wide association study. Mol. Psychiatry 15(7), 756-766 (2010).
- Bertram L, Tanzi RE. Genome-wide association studies in Alzheimer’s disease. Human. Mol. Genetics 18(R2), (2009).
- Tosto G, Reitz C. Genome-wide association studies in Alzheimer’s disease: a review. Curr. Neurol. Neurosci. Rep 13(10), 381 (2013).
- Shen L, Jia J. An Overview of Genome-Wide Association Studies in Alzheimer’s disease. Neurosci. Bulletin 32(2), 183-190 (2016).
- Grande I, Berk M, Birmaher B, et al. Bipolar disorder. Lancet 387(10027), 1561-1572 (2016).
- Schmitt A, Malchow B, Hasan A, et al. The impact of environmental factors in severe psychiatric disorders. Front. Neurosci 8(1), 19 (2014).
- Reich T, Clayton PJ, Winokur G. Family history studies: V.The genetics of mania. Am. J. Psychiatry 125(10), 1358-1369(1969).
- Gao J, Jia M, Qiao D, et al. TPH2 gene polymorphisms and bipolar disorder: A meta-analysis. Neuropsychiatric. Genetics 171(2), 145-152 (2016).
- Hou L, Bergen SE, Akula N, et al. Genome-wide association study of 40,000 individuals identifies two novel loci associated with bipolar disorder. Human Mol. Genetics 25(15), 3383-3394 (2016).
- Baum AE, Akula N, Cabanero M, et al. A genome-wide association study implicates diacylglycerol kinase eta (DGKH) and several other genes in the etiology of bipolar disorder. Mol. Psychiatry 13(2), 197-207 (2008).
- Ferreira MAR, O’Donovan MC, Meng YA, et al. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat. Genetics 40(9), 1056-1058 (2008).
- Chen DT, Jiang X, Akula N, et al. Genome-wide association study meta-analysis of European and Asian-ancestry samples identifies three novel loci associated with bipolar disorder. Mol. Psychiatry 18(2), 195-205 (2013).
- Sklar P, Ripke S, Scott LJ, et al. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat. genetics 43(10), 977 (2011).
- Cross-Disorder Group of the Psychiatric Genomics Consortium. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet 381(9875), 1371-1379 (2013).
- Ripke S, Wray NR, Lewis CM, et al. A mega-analysis of genome-wide association studies for major depressive disorder. Mol. Psychiatry 18(4), 497 (2013).
- Mühleisen TW, Leber M, Schulze TG, et al. Genome-wide association study reveals two new risk loci for bipolar disorder. Nat. Comm 5(1), (2014).
- Soronen P, Ollila HM, Antila M, et al. Replication of GWAS of bipolar disorder: Association of SNPs near CDH7 with bipolar disorder and visual processing. Mol. Psychiatry 15(1), 4-6 (2010).
- Dima D, de Jong S, Breen G, et al. The polygenic risk for bipolar disorder influences brain regional function relating to visual and default state processing of emotional information. NeuroImage. Cli 12(1), 838-844 (2016).
- Ikeda M, Takahashi A, Kamatani Y, et al. A genome-wide association study identifies two novel susceptibility loci and trans population polygenicity associated with bipolar disorder. Mol. Psychiatry 23(3), 639-647 (2017).
- Higgins GA, Allyn-Feuer A, Barbour E, et al. A glutamatergic network mediates lithium response in bipolar disorder as defined by epigenome pathway analysis. Pharmacogenomics 16(14), 1547-1563 (2015).
- Sprich S, Biederman J, Crawford Mh, et al. Adoptive and Biological Families of Children and Adolescents With ADHD. J. Am. Acad. Child. Adolesc. Psychiatry 39(11), 1432-1437 (2000).
- Cantwell DP. Genetics of hyperactivity. J. Child. Psychology. Psychiatry 16(3), 261-264 (1975).
- Demontis D, Walters RK, Martin J, et al. Discovery of the first genome-wide significant risk loci for ADHD. bioRxiv 145581 (2017).
- Stergiakouli E, Martin J, Hamshere ML, et al. Association between polygenic risk scores for attention-deficit hyperactivity disorder and educational and cognitive outcomes in the general population. Int. J. Epidemiol 46(2), 421-428 (2016).
- Ritter ML, Guo W, Samuels JF, et al. Genome Wide Association Study (GWAS) between Attention Deficit Hyperactivity Disorder (ADHD) and Obsessive Compulsive Disorder (OCD). Frontiers. Mol. Neurosci 10(1), (2017).
- Ashmore K, Cheng F. Genome-wide association studies on Attention Deficit Hyperactivity Disorder. Clin. Exp. Pharmacol (2013).
- Zayats T, Athanasiu L, Sonderby I, et al. Genome-wide analysis of attention deficit hyperactivity disorder in Norway. PLoSONE 10(4) (2015).
- Siddique N, Siddique T. Genetics of Amyotrophic Lateral Sclerosis. Phys. Med. Rehabil. Clin. N. Am 19(3), 429-439 (2008).
- Corcia P, Couratier P, Blasco H, et al. Genetics of amyotrophic lateral sclerosis. Rev. Neurol 173(5), 254-262 (2017).
- Lattante S, Ciura S, Rouleau GA, et al. Defining the genetic connection linking amyotrophic lateral sclerosis (ALS) with frontotemporal dementia (FTD). Trends. Genetics 31(5), 263-273 (2015).
- Ryan RM, Deci EL. On Happiness and Human Potentials: A Review of Research on Hedonic and Eudaimonic Well-Being. Ann. Review. Psychology 52(1), 141-166 (2001).
- Fredrickson BL, Grewen KM, Coffey KA, et al. A functional genomic perspective on human well-being. Proceedings of the National Academy of Sciences. 110(33), 13684-13689 (2013).
- Brown NJL, MacDonald DA, Samanta MP, et al. A critical reanalysis of the relationship between genomics and well-being. Proceedings of the National Academy of Sciences. 111(35), 12705-12709 (2014).
- Ploug T, Holm S. Informed consent and registry-based research - The case of the Danish circumcision registry. BMC. Med. Ethics 18(1), (2017).
- Haug CJ. Whose Data Are They Anyway? Can a Patient Perspective Advance the Data-Sharing Debate?. New. Eng. J. Med 376(23), 2203-2205 (2017).
- Alirol E, Kuesel AC, Guraiib MM, et al. Ethics review of studies during public health emergencies - the experience of the WHO ethics review committee during the Ebola virus disease epidemic. BMC. Med. Ethics 18(1), 43 (2017).
- Rahimzadeh V, Bartlett G. Genetics and primary care: Where are we headed? J. Transl. Med 12(1), (2014).
- Hurlimann T, Groisman IJ, Godard B. The elusive ideal of inclusiveness: lessons from a worldwide survey of neurologists on the ethical issues raised by whole-genome sequencing. BMC. Med. Ethics 18(1), (2017).
- Caruso JP, Sheehan JP. Psychosurgery, ethics, and media: a history of Walter Freeman and the lobotomy. Neurosurgical Focus 43(3), E6 (2017).
- Borry P, Bentzen HB, Budin-Ljøsne I, et al. The challenges of the expanded availability of genomic information: an agenda-setting paper. J. Comm. Genetics. 9(2), 1-14 (2017).
- Haga SB, Friedman B, Richard G. Considering the Benefits and Risks of Research Participants' Access to Sequence Data. Genet. Test. Mol. Biomarkers 21(12), 717-721 (2017).
- Wouters RHP, Cornelis C, Newson AJ, et al. Scanning the body, sequencing the genome: Dealing with unsolicited findings. Bioethics 31(9), 648-656 (2017).
- Groisman IJ, Mathieu G, Godard B. Use of next generation sequencing technologies in research and beyond: Are participants with mental health disorders fully protected? BMC. Med. Ethics 13(1), (2012).
- Santosh P, Arora N, Sarma P, et al. Interaction map and selection of microRNA targets in Parkinson's disease-related genes. BioMed. Res. Int 2009, 363145 (2009).
- Shinde S, Arora N, Bhadra U. A Complex Network of MicroRNAs Expressed in Brain and Genes Associated with Amyotrophic Lateral Sclerosis. Int. J. Genomics 2013, 383024 (2013).
- Choudhury A, Sahu T, Ramanujam PL, et al. Neurochemicals, behaviours and psychiatric perspectives of neurological diseases. Neuropsychiatry 8(1), 395-424 (2018).