Review Article - Imaging in Medicine (2013) Volume 5, Issue 1
Noninvasive monitoring of liver cell transplantation
Nathanael Raschzok*1, M Haluk Morgül2, Lars Stelter3 & Igor M Sauer11General, Visceral & Transplant Surgery, Experimental Surgery & Regenerative Medicine, Campus Virchow Klinikum, Charité – Universitätsmedizin Berlin, Germany
2Department of Visceral, Transplantation, Thoracic & Vascular Surgery, Universitätsklinikum Leipzig, Germany
3Department of Radiology, Campus Virchow Klinikum, Charité – Universitätsmedizin Berlin, Germany
- Corresponding Author:
- Nathanael Raschzok
General, Visceral & Transplant Surgery
Experimental Surgery & Regenerative Medicine
Campus Virchow Klinikum, Charité – Universitätsmedizin Berlin, Germany
Tel: +49 30 450 652 356
Fax: +49 30 450 559 987
E-mail: nathanael.raschzok@charite.de
Abstract
Keywords
cell tracking ▪ liver cell transplantation ▪ MPIO ▪ MRI ▪ radionuclide imaging ▪ SPIO
Liver cell transplantation: conceptual background, current clinical situation & the need for noninvasive monitoring
Liver cell transplantation (LCT) is an evolving therapeutic approach for the treatment of a variety of liver diseases and is a potential future alternative to solid liver transplantation [1]. LCT is based on the administration of liver cells in suspension, which implies several conceptual advantages over whole-organ transplantation. Cells are isolated from donor livers or liver lobes that were rejected or unused for whole-organ transplantation, and this expands the donor pool for liver grafts and allows for the treatment of multiple patients with cells from one donor organ. Liver cells can be cryopreserved prior to transplantation, which enables the pooling of cells and on-demand or scheduled applications. Cells can be infused either into the liver or to an ectopic implantation site, such as the spleen or the peritoneal cavity, by interventional procedures. This method is also less invasive than whole-organ transplantation and offers the chance to treat critically ill or very young patients who are not suitable for whole-organ transplantation. The cells can engraft in the recipient liver or at the ectopic implantation site and provide metabolic activity without the need for removing the diseased liver. Thus, the native liver can be left in place and may have a chance of recovery [2–4].
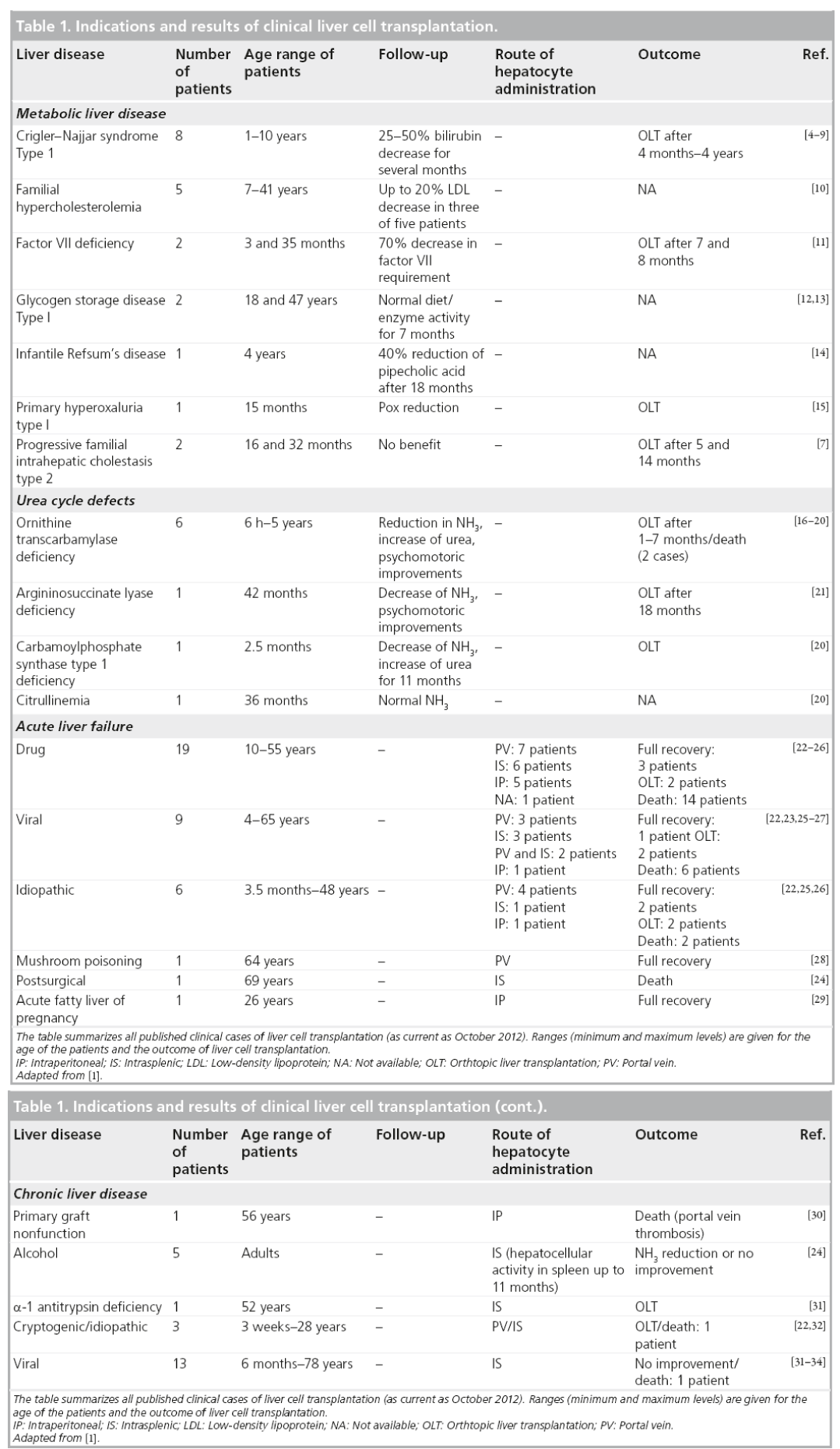
Following extensive animal studies, LCT has been clinically evaluated for the major indications of liver transplantation (Table 1) [4–34]. Unfortunately, LCT has not yet achieved sustainable benefits for patients with acute liver failure and chronic liver disease [1]. However, LCT has evolved as an effective bridging strategy for patients suffering from inborn metabolic liver disorders. In cases with a single deficient enzyme, such as urea cycle defects [16–21] or glycogen storage disease [12,13], donor hepatocytes can substitute the missing function without the need for replacing the whole organ. Approximately 35 children and adults who received LCT for liver-based metabolic disease have been reported in the literature to date. The majority of these studies were conducted within the last few years [35]. Liver cell administration proved to be feasible and safe, and the majority of the cases demonstrated temporal clinical improvements, such as bilirubin or urea reduction [20] or the presence of soluble factors synthesized by transplanted cells [19]. However, in almost all cases, patients needed to undergo liver transplantation a few months after LCT due to recurrence of symptoms and further aggravation of the disease.
There are still many open questions and barriers for the successful treatment of liver disease by LCT [36]. One of the persisting problems is the lack of evidence for the longterm engraftment and function of transplanted cells. Possible reasons for the unsustainable longterm outcome of LCT include insufficient cell translocation through the endothelial cell barrier [37], insufficient cell engraftment into the liver plates [38] and destruction of the engrafted cells by the immune system [39,40]. Unfortunately, little is known about the processes that occur during and after LCT in humans. Another major challenge is that primary human hepatocytes are currently the only source for clinical LCT. With a continuously decreasing number of available organs for hepatocyte isolation, there is a critical scarcity of hepatocytes for transplantation. Strategies to overcome this issue include the use of stem cells or induced pluripotent stem cells to generate hepatocytes. Protocols for hepatocyte generation have already been developed, but the safety and clinical applicability of such cells has not yet been proven [41,42].
The optimization of clinical LTC has mainly been obstructed by the inability to track the fate of the transplanted liver cells in patients once they have been infused. Conventional clinical imaging techniques, such as ultrasound or CT, are not suited to visualize transplanted cells. Clinical evaluation of transplanted cell activity mainly relies on the measurement of soluble factors or enzymatic activity provided by the graft [21]. This approach is suitable in cases of metabolic disorders, when single enzymes are missing and the metabolic activity of transplanted cells can be distinguished from the patient’s own liver cells. However, the monitoring of enzymatic activities does not enable localization of the transplanted cells. Another clinical option for evaluating liver cell grafts is histological analysis of tissue samples obtained through biopsies of the engraftment organ [43]. Immunohistological techniques, such as f luorescence in situ hybridization for the Y chromosome sequences in cases of sex-mismatched transplantations or human leukocyte antigen class I tissue typing, enable the detection of transplanted cells in biopsies of the recipient liver. However, biopsies have intrinsic risks for the patient and cannot be performed repetitively. Moreover, real-time anatomical localization of transplanted cells in the recipient organ is not possible.
Noninvasive monitoring of LCT by means of tracking transplanted cells with imagingbased approaches is the ultimate goal and, presumably, the best solution to address the questions of insufficient engraftment and the long-term function of transplanted liver cells. Such approaches should ideally involve repetitive imaging of high sensitivity and spatial resolution, making continuous visualization and tracking of transplanted liver cells possible. Experimental and initial clinical studies have investigated various imaging-based approaches for the noninvasive monitoring of LCT. This article summarizes and critically discusses these approaches, and provides an outlook on possible clinical applications for the near future.
Radionuclide-based imaging techniques for the visualization of transplanted liver cells
Nuclear imaging techniques rely on the properties of radioactive substances, combined with pharmaceutical compounds or elements, to emit radiation based on nuclear decay. Upon administration to the patient, the electromagnetic radiation emitted can be captured by external detectors (γ-cameras) and, after digital reconstruction, can be used to generate images based on specific metabolic processes [44,45]. Nuclear imaging techniques provide highly selective information in vivo as the different tissues in the living body have differential internalization of the specific radiopharmaceuticals. Therefore, nuclear imaging modalities, such as scintigraphy, single photon emission CT (SPECT) and PET are commonly used in clinical practice as valuable diagnostic tools in oncologic and inflammatory diseases. In addition to their high sensitivity, radionuclide-imaging modalities can provide additional information regarding the metabolism through functional imaging, which makes this modality highly attractive for noninvasive monitoring of LCT [46]. For imaging purposes, donor cells can be labeled with radionuclides both prior to and after administration [47–49]. In vitro labeling enables highly selective imaging of the transplanted cells. In vivo radionuclide labeling is based on specific hepatocellular receptor expression, for example the hepatocyte asialoglycoprotein receptor, which enables selective uptake of intravenously administered radionuclides such as technetium-99m (99mTc) -labeled galactosyl-serum albumin [50,51]. However, in order to visualize and localize the cells later, the cells of interest must demonstrate different radionuclide uptake characteristics than the surrounding cells.
Both in vitro and in vivo radionuclide labeling approaches have already been used clinically for the noninvasive monitoring of transplanted liver cells. The very first in-human LCT, described by Mito et al. in 1992, used conventional scintigraphy and 99mTc-labeled radionuclides to visualize the transplanted liver cells. In this first study, autologous hepatocytes were isolated from surgically resected liver tissue and transplanted to the spleen of patients with chronic liver failure [33]. As hepatocytes demonstrate a different 99mTc internalization capacity compared with resident cells in the spleen, heterotopic transplantation enabled long-term visualization of hepatocytes in the spleen for up to 11 months following transplantation by repeated administration of the radioactive compound. Sterling and Fisher used the same approach in a patient with chronic liver disease. They infused allogenic hepatocytes via the splenic artery and demonstrated hepatocellular activity in the spleen at 9 and 23 days following LCT [34]. By contrast, Bohnen et al. used an in vitro labeling approach for radionuclide imaging of transplanted liver cells [52]. Following labeling with indium-111 (111In), transplanted cells could be detected in the liver of the recipient over a period of 48 h by scintigraphy.
The obvious limitation of scintigraphy is the very coarse anatomical information since a planar scintigram provides only 2D information. Tomographic nuclear medicine imaging methods, such as PET and SPECT or, when combined with CT, PET-CT and SPECT-CT, are more promising approaches for in-depth localization of transplanted cells. However, based on current literature, there has not been clinical use of these tools in LCT until now.
A general limitation for nuclear-based imaging techniques is the short physical half-life of the clinically used radionuclides (99mTc: 6 h and 111In: 2.8 days) [53]. The application of radioactive tracers with a longer half-life could overcome this problem, but translation into clinical LCT may be limited based on radiation hygiene requirements. Moreover, the renal elimination and, most importantly, the chemical stability of the radioactive compound need to be carefully evaluated to avoid data misinterpretation as the mononuclear phagocytic system could internalize the unbound radionuclide nonspecifically [54,55].
In most of the clinical cases of LCT, hepatocytes were transplanted via the portovenous system and were expected to engraft in the recipient liver. Thus, in vivo radionuclide labeling of the donor cells following orthotopic LCT would be hampered unless the uptake characteristics of the donor cells could be manipulated to differ from the hosts’ liver cells. Unfortunately, genetic manipulation of the graft by using, for example, reporter gene-mediated imaging is still in preclinical investigation and cannot yet be transferred into a clinical setting [56–58].
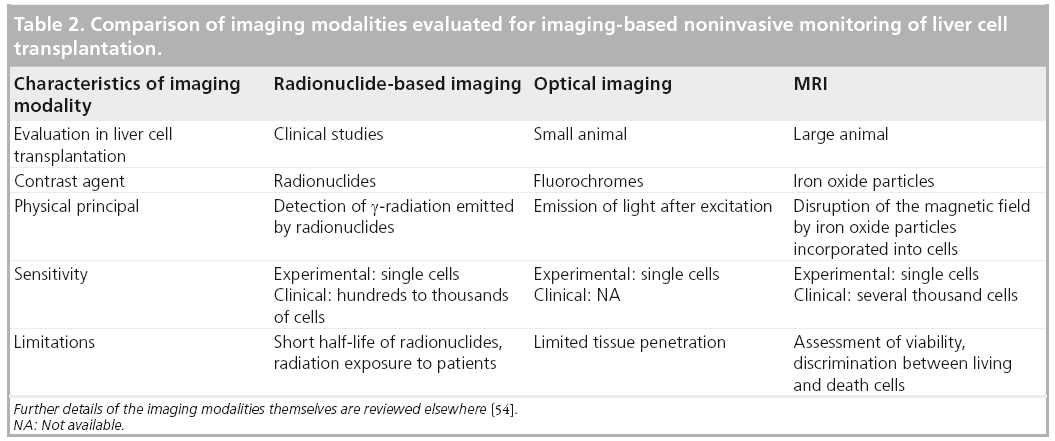
In conclusion, radionuclide-based imaging has already been clinically used to noninvasively monitor LCT. Based on the current knowledge, nuclear imaging-based approaches are useful for the short-term noninvasive analysis of the biodistribution and function of transplanted hepatocytes, and for noninvasive monitoring of hepatocytes transplanted into ectopic implantation sites. Further studies are necessary in order to evaluate the diagnostic advantages of tomographic methods for radionuclide imaging of LCT.
MRI enables noninvasive tracking of transplanted liver cells
MRI: concept & background
MRI is a radiation-free imaging technique routinely available in most hospitals. MRI provides high spatial resolution and excellent soft tissue contrast. Imaging can be repeated without harming the patient, making this technology highly attractive for the long-term tracking of transplanted liver cells [59,60]. MRI uses the property of NMR to image the nuclei of hydrogen atoms inside the body. In brief, a magnetic field aligns the protons of the hydrogen nuclei. Radio frequency pulses are used to alter the alignment of this magnetization, which causes the nuclei to produce a rotating magnetic field detectable by an MRI scanner. Different variables, such as the spin density, T1 and T2 relaxation times, and flow and spectral shifts, are used to construct images of the scanned area of the body [61,62]. MRI resolution depends on the field strength (clinical MRI: 1.0–3.0 Tesla; experimental MRI: up to 17 Tesla) and the size of the coil used for imaging. A decrease in field strength or an increase in the size of the coil leads to a reduction in the sensitivity, contrast and resolution. Furthermore, the region of the body that is investigated plays an important role in the image quality. For example, T2* sequences can be used to generate high-quality images of the brain, while in the abdomen, only low quality can be achieved due to moving artifacts by the bowel [63]. Figure 1 provides an overview of the workflow that would be necessary for clinical MRI monitoring of LCT.
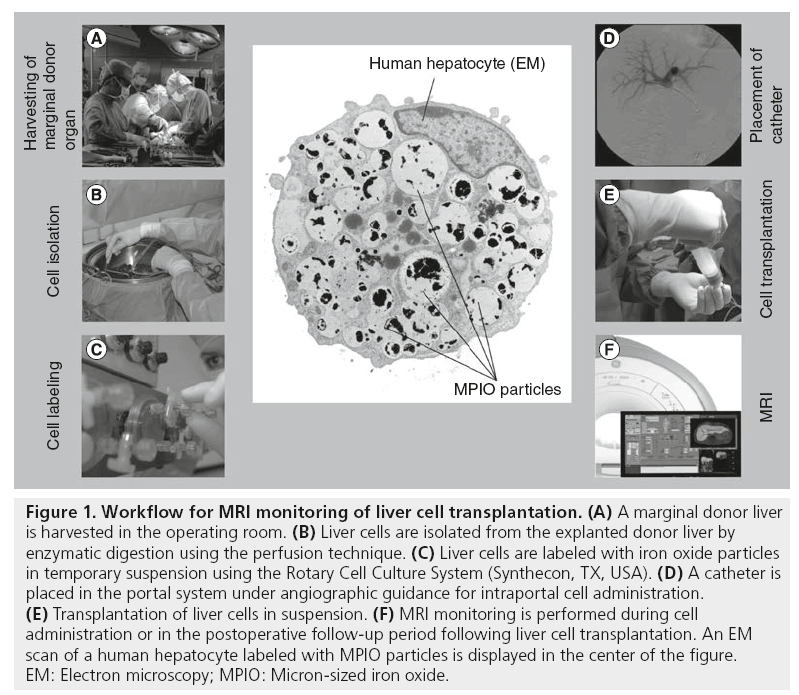
Figure 1: Workflow for MRI monitoring of liver cell transplantation. (A) A marginal donor liver is harvested in the operating room. (B) Liver cells are isolated from the explanted donor liver by enzymatic digestion using the perfusion technique. (C) Liver cells are labeled with iron oxide particles in temporary suspension using the Rotary Cell Culture System (Synthecon, TX, USA). (D) A catheter is placed in the portal system under angiographic guidance for intraportal cell administration. (E) Transplantation of liver cells in suspension. (F) MRI monitoring is performed during cell administration or in the postoperative follow-up period following liver cell transplantation. An EM scan of a human hepatocyte labeled with MPIO particles is displayed in the center of the figure. EM: Electron microscopy; MPIO: Micron-sized iron oxide.
Iron oxide particles: intracellular contrast agents for cell detection by MRI
Intracellular contrast agents are necessary to enable detection of transplanted cells by MRI. MRI contrast agents are generally based on paramagnetic or superparamagnetic substances. Since the superparamagnetic agents, such as iron oxide particles, provide stronger contrast than the paramagnetic agents, these substances are generally preferred for cellular MRI [64,65]. When superparamagnetic particles are placed in an external magnetic field, the magnetic dipoles orient to produce local disruptions in the magnetic field. MRI protocols that are sensitive to the dephasing effect of superparamagnetic particles are used to visualize particle-labeled cells. Using T2- or T2*-weighted sequences, particle-labeled cells appear as signal extinctions or signal voids [66]. Superparamagnetic iron oxide (SPIOs) particles consist of an iron core covered with an appropriate encapsulation material that prevents iron toxicity. Iron oxide particles are available in different sizes. Sizes range from nanometers (ultrasmall SPIO: size <50 nm; SPIOs: size >50 nm) to micrometers (micron-sized iron oxide [MPIO] particles).
Protocols for labeling hepatocytes with SPIO particles
Nanometer-sized ferumoxides (USA: Feridex®/ Europe: Endorem®) with a particle size of 50–180 nm are commercially available and clinically approved for MRI of the liver. Feridex/ Endorem particles consist of SPIO nanoparticles coated with dextran. Cellular labeling with SPIOs can be achieved through the simple incubation of cultivated cells with the particles. Based on experiences with other cell types, a general protocol for labeling hepatocytes with SPIOs was published by Modo et al. [67]. Puppi et al. evaluated the labeling of human hepatocytes in adhesion culture with Endorem SPIOs using incubation concentrations ranging from 0 to 150 μg of Fe/ ml and incubation times from 2 to 24 h [68]. They demonstrated a time- and dose-dependent increase in the intracellular iron concentrations after incubation with SPIOs up to 18.5 ± 3.4 pg of Fe/cell. The authors used a similar protocol (16 h of incubation and 50 μg of Fe/ml) for labeling primary human hepatocytes with Endorem SPIOs and achieved an iron load of 21.4 ± 13.2 pg of Fe/ cell [69]. In both studies, SPIO labeling produced only minimal adverse effects on the metabolic activity of hepatocytes. Urea synthesis of primary human hepatocytes was not affected by SPIO labeling over a period of up to 14 days in both studies. Puppi et al. observed a dose-dependent decrease in albumin production at day 4 after labeling with subsequent recovery in cells labeled with >100 μg Fe/ml. Moreover, they found a significant decrease in albumin production after 24 h of labeling with SPIOs compared with 16 h labeling at incubation concentrations >25 μg Fe/ml [68]. Transferrin synthesis and CYP1A1/2 activity of SPIO-labeled cells were similar to unlabeled cells. However, the authors observed increased oxygen species formation in human hepatocytes following SPIO labeling [69]. Overnight incubation with SPIOs is a rather long incubation time for hepatocytes in culture as the efficiency of resuspending hepatocytes for transplantation is dramatically decreased over time. As is known from other cellular systems, particle uptake can be enhanced using transfection agents or using particle surface modifications [70]. The authors modified MagForce® (MagForce Applications GmbH, Berlin, Germany) SPIO particles, which are aminosilane-coated iron oxide nanoparticles (mean size of ~100 nm), with the HIV membrane-translocating Tat peptide [71]. This approach enabled sufficient hepatocyte labeling within a 1 h incubation time. An alternative is the use of an electrostatic interaction to enhance the particle internalization process. For example, Luciani et al. achieved sufficient mouse hepatocyte labeling by 15 min of incubation with anionic iron oxide ultrasmall SPIO [72].
The detectability of labeled cells in MRI depends on the number of particles that are incorporated into the cells and the number of cells that are visualized. Puppi et al. demonstrated a positive correlation between the number of SPIO-labeled hepatocytes in an agarosis phantom model and the change in the T2 relaxivity using an experimental 7.0 Tesla MR system [68]. At 24 h following labeling, concentrations of 2000, 4000 and 20,000 cells/μl labeled with 50 μg of Fe/ml for 16 h induced a decrease of 14, 48 and 84%, respectively, on T2 relaxivity compared with nonlabeled cells. The authors used a clinical 3.0 Tesla MRI scanner to investigate the detectability of SPIO-labeled cells [69]. Evaluation of the detectability threshold was performed morphometrically by clinical and experimental radiologists. Based on the authors evaluation, Endorem SPIO-labeled human hepatocytes were detectable down to a minimum number of 250,000 cells on T1-weighted MRI, and down to a number of 50,000 cells on T2*-weighted MRI [69].
MPIO: ‘sizing it up’
MPIOs have a significantly larger iron core compared with SPIOs and thereby dramatically enhance the detectability of labeled cells by MRI [66]. Shapiro et al. were the first to investigate MPIO labeling (diameter: 0.96–5.8 μm) with murine hepatocytes [73]. They demonstrated the feasibility of detecting single MPIO-labeled hepatocytes in vitro using an experimental MR scanner with field strengths of 7.0 and 11.7 Tesla. Raschzok et al. evaluated the labeling of primary human hepatocytes in adhesion culture with 1.6 μm MPIOs [74]. An uptake of approximately 18 MPIOs per cell (iron content: ~18 pg iron/ cell) and almost 100% labeling efficiency following 4 h of incubation was achieved. Using a T2*-weighted sequence and a 3 cm surface coil, MPIO-labeled human hepatocytes were detectable on single-cell level by 3.0 Tesla MRI. Measurements were also performed using a wholebody coil and sequences applicable for clinical abdominal MRI [69]. Under these conditions, MPIO-labeled cells induced clearly detectable signal extinctions from at least 50,000 labeled cells on T1-weighted MRI. To prepare MPIOlabeled hepatocytes ready for transplantation without the need for resuspension from culture plates prior to transplantation, Kammer et al. developed a protocol for hepatocyte labeling in suspension using the Rotary Cell Culture System (Synthecon, TX, USA) [75]. No adverse effects of MPIO-labeling in adhesion or in rotation on the metabolic activity of human hepatocytes were found. Moreover, MPIOs did not lead to reactive oxygen species formation of labeled hepatocytes, even at a similar iron load compared with SPIO-labeled hepatocytes [69]. However, the commercially available MPIOs (Bangs Laboratories, Inc.™, IN, USA) are not clinically applicable since a styrene–divinyl benzene polymer shell covers the iron core. Nkansah et al. recently reported on the fabrication of MPIOs composed of poly(lactide-co-glycolide) and cellulose, which are both US FDA-approved polymers [76]. In vitro experiments with human adenocarcinoma cells and mouse mesenchymal stem cells (MSCs) demonstrated no cytotoxicity of these particles. The authors recently reported on the development of functionalizable silica-based MPIOs, which also consist of biocompatible materials and should be easily translated into clinical applications of cellular MRI as our studies demonstrated no adverse effects of silica-based MPIOS on human and rat hepatocytes [77].
Small animal studies on the MRI tracking of transplanted liver cells
While various studies have been published on the MRI tracking of particle-labeled stem cells transplanted into the liver, only a few studies are available on the tracking of transplanted liver cells by MRI. Ju et al. were the first to evaluate MRI tracking of stem cells in the liver [78]. They used custom-made Fe2O3–poly-llysine superparamagnetic particles to label rodent bone MSCs. Cells were transplanted via intrasplenic injection, which is a common route of administration for hepatocyte transplantation in rodents. Animals were pretreated with carbon tetrachloride in order to induce liver damage and subsequent cirrhosis. Imaging was performed using a 1.5 Tesla MR scanner with a 12.7 cm receiver surface coil. Upon injection of particle labeled bone MSCs, the contrast-to-noise ratios on T2*-weighted images of the liver decreased significantly and returned to normal levels within 14 days. Transplantation of unlabeled cells did not lead to a signal decrease in the liver. Bos et al. obtained similar results using MSCs labeled with Endorem SPIO transplanted into the liver of animals pretreated with carbon tetrachloride [79]. The cells were detected for up to 12 days in the liver using a 1.5 Tesla MR scanner and a T2*-weighted gradient-echo MR sequence. Signal intensity loss and fading over time could be confirmed with serial R2* mapping. At the time of histologic analysis, the signal intensity loss correlated with iron-loaded MSCs in hepatic sinusoids. Both studies demonstrated the feasibility of monitoring SPIO-labeled stem cells in the liver of living animals by MRI, providing a basis for further studies with particle-labeled hepatocytes.
Luciani et al. performed in vivo studies using mouse hepatocytes labeled with anionic iron oxide ultrasmall SPIO [72]. They used a standard 1.5 Tesla magnet and imaging protocols similar to human applications. Labeled hepatocytes were injected via the portal vein into normal mice. Following the administration of labeled cells, recipient livers demonstrated well-defined nodular foci of low-signal intensity on MRI. In parallel, a significant decrease in both the liver T2 values and liver-to-muscle relaxivity index was observed, and these continued decreasing until 8 days post-transplantation. Histological analysis with Prussian blue staining highlighted the presence of iron oxide particle-labeled hepatocytes within the liver parenchyma. The clusters of Prussian bluepositive hepatocytes were distributed throughout the liver and correlated with the foci of low-signal intensity detected on MRI.
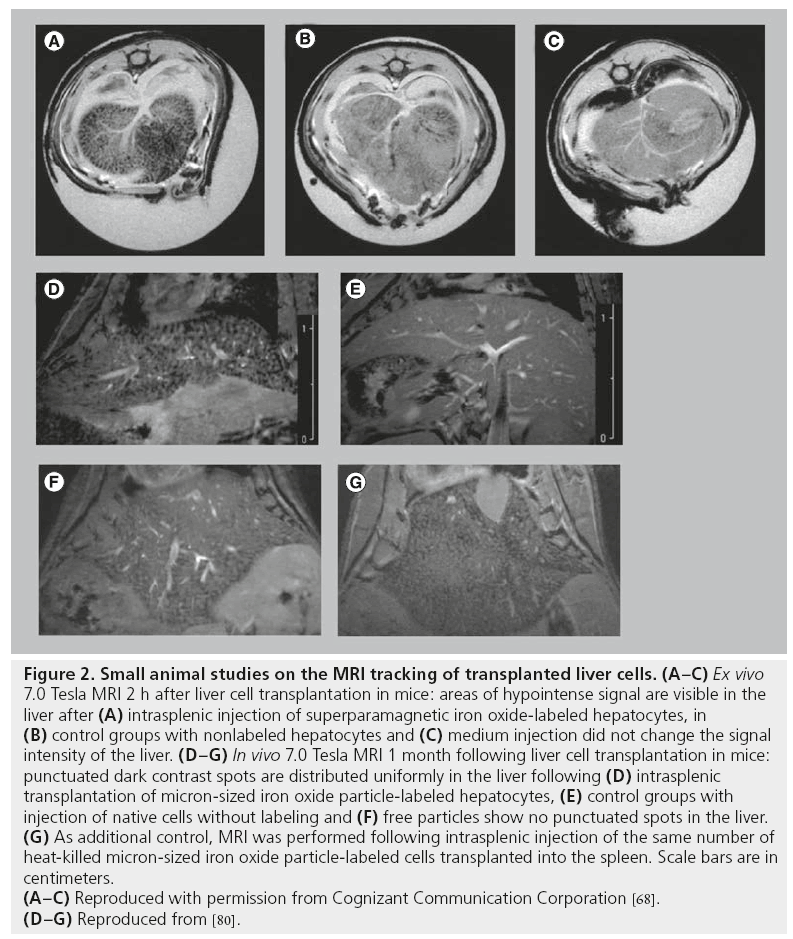
Puppi et al. tested whether SPIO-labeled human hepatocytes can be detected following intrasplenic transplantation into immunodeficient mice [68]. Imaging was performed 2 h after hepatocyte injection and after sacrifice of the animals. A 7.0 Tesla MR scanner and T2*- weighted sequences were used for imaging. Under these conditions, labeled cells clearly caused a decrease in the signal intensity in the liver, while nonlabeled cells did not produce signal changes (Figure 2A–C). However, imaging was performed ex vivo, which guaranteed optimized conditions for MRI but is not directly comparable with in vivo MRI, where motion artifacts can decrease image quality and resolution.
Figure 2: Small animal studies on the MRI tracking of transplanted liver cells. (A–C) Ex vivo
7.0 Tesla MRI 2 h after liver cell transplantation in mice: areas of hypointense signal are visible in the
liver after (A) intrasplenic injection of superparamagnetic iron oxide-labeled hepatocytes, in
(B) control groups with nonlabeled hepatocytes and (C) medium injection did not change the signal
intensity of the liver. (D–G) In vivo 7.0 Tesla MRI 1 month following liver cell transplantation in mice:
punctuated dark contrast spots are distributed uniformly in the liver following (D) intrasplenic
transplantation of micron-sized iron oxide particle-labeled hepatocytes, (E) control groups with
injection of native cells without labeling and (F) free particles show no punctuated spots in the liver.
(G) As additional control, MRI was performed following intrasplenic injection of the same number of
heat-killed micron-sized iron oxide particle-labeled cells transplanted into the spleen. Scale bars are in
centimeters.
(A–C) Reproduced with permission from Cognizant Communication Corporation [68].
(D–G) Reproduced from [80].
Shapiro et al. investigated in vivo MRI tracking of MPIO-labeled hepatocytes in a mouse model [80]. Imaging was performed with T2*-weighted multislice gradient-echo sequences using a 72 mm volume coil in conjunction with a 35 mm surface receive-only coil and a 7.0 Tesla MRI system. MPIO-labeled hepatocytes were detectable as punctuated, dark contrast regions in the liver of recipients 4 weeks after intrasplenic injection (Figure 2D). MRI of perfused, fixed samples in combination with histological evaluation was used to confirm the presence of dispersed single hepatocytes grafted into the livers. Control groups with free particles and heat-killed MPIO-labeled cells were used to demonstrate that the contrast was due to dispersed, single MPIO-labeled cells (Figure 2E–G). Shapiro et al. proved the feasibility of detecting single MPIO-labeled hepatocytes in vivo [80]. However, when comparing their data with results obtained from other studies, it is important to consider that they used high-field MRI, which guaranteed high-image resolution and high contrast by particle-labeled cells.
Leconte et al. performed imaging studies with 1.6 μm of MPIO-labeled hepatocytes transplanted in mice using a 1.5 Tesla MR scanner [81]. The aim of their study was to investigate whether MRI can be used to study the transport of cells to the liver, the intrahepatic distribution and the engraftment of labeled hepatocytes. Therefore, they compared hepatocyte transplantation in untreated animals versus animals treated with cyclophosphamide, which disrupts the hepatic sinusoidal endothelium of the liver sinusoids and should promote hepatocyte engraftment. They observed significantly lower signal intensity in the liver of rats pretreated with cyclophosphamide compared with untreated rats at 7 days following MPIOlabeled hepatocyte transplantation. At the time of histology, more particles were found in pretreated animals compared with the untreated animals; however, the majority of the MPIO were located within Kupffer cells and not within hepatocytes. This observation was not surprising since it is known that 70–85% of transplanted hepatocytes die in the sinusoids within the first 24–48 h after transplantation, and are taken up by the Kupffer cells [35]. However, these results demonstrated that MRI with MPIOlabeled cells is limited to a short period of time as particles taken up by macrophages cannot be discriminated from particles in living cells. Moreover, MR images of the liver do not reflect the exact location of the surviving hepatocytes since most iron oxide particles were not located within hepatocytes, but were instead within Kupffer cells. Importantly, their results were contrasted with those of Shaprio et al., who could discriminate MR signals obtained by labeled living cells and labeled dead cells or free particles [80]. Presumably, a MR scanner with a stronger magnetic field would have been necessary to distinguish the signal extinctions induced by MPIO-labeled hepatocytes from Kupffer cells having internalized free particles.
In conclusion, small animal studies proved the general feasibility of MRI-based monitoring of LCT. Both SPIO and MPIO are suitable as contrast agents for hepatocyte labeling and induce sufficient contrast for MRI. When comparing the results of these studies, technical factors, such as the MR system and the imaging sequences, as well as the type of particle used for cellular labeling, must be taken into account. Moreover, these results demonstrated that cellular MRI is limited to a distinct period of time and that issues arise with monitoring cellular survival.
Large animal studies on the MRI tracking of transplanted liver cells
Large animal studies are crucial prior to clinical translation of an experimental imaging approach. In particular, in the field of MRI, where image resolution depends on various technical factors, large animal studies are mandatory for investigating the possible clinical relevance of an imaging modality. To the best of our knowledge, three studies have been published addressing MRI monitoring of liver or stem cell transplantation to the liver in a large animal model.
Shi et al. investigated MRI tracking of MSCs after intraportal transplantation in a swine model of acute liver injury (d-galactosamine poisoning) [82]. MSCs were labeled with Feridex SPIO and poly-l-lysine was used as a transfection agent to enhance the labeling efficiency. Imaging was performed at 3, 7 and 14 days after cell injection with a 1.5 Tesla imaging device. They observed a decrease in the signal intensity of the liver 6 h after intrahepatic transplantation of labeled MSCs on T2*-weighted MRI. Thereafter, the signal intensity gradually reached normal levels. Visible differences were not observed following injection of nonlabeled cells. These results were comparable with the results obtained from small animal models with MRI tracking of SPIO-labeled cells, showing that infusion of SPIO-labeled cells leads to a visible decrease in the signal intensity of the liver, and this signal intensity returns to normal levels within a distinct period to time.
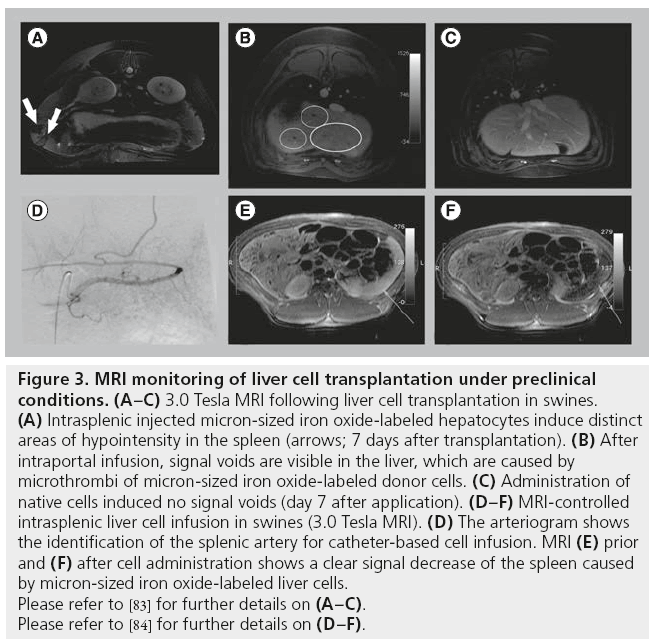
The authors investigated MRI monitoring of transplanted liver cells in preclinical swine models. In a first study, MPIO-labeled liver cells were transplanted via intraportal infusion into the liver, via direct injection into the splenic parenchyma, or via intra-arterial infusion to the spleen of nonpretreated animals [83]. Imaging was performed using a clinical 3.0 Tesla MR scanner and a 30 cm whole-body coil. The T2*-weighted sequence, which previously yielded the best results for imaging particle-labeled cells, was not applicable in our setting because of the lengthy breath hold periods needed for abdominal MRI at 3.0 Tesla. Using a 3D T1-weighted liver acquisition with volume acceleration sequence, distinct areas of hypointensity were found in the spleen following MPIO-labeled hepatocyte injection (Figure 3A). These areas of hypointensity could be correlated with the engrafted hepatocytes by immunohistology and electron microscopy. However, injection of pure particles led to similar hypointense areas in the spleen, indicating that discrimination between labeled cells and free particles is not possible. Following intraportal infusion, signal voids appeared in the liver and were histologically correlated to microthrombi of MPIO-labeled cells in distal portal branches. Signal voids were not found following transplantation of unlabeled cells or injection of pure particles (Figure 3B & C). Microembolization is a common unwanted event following LCT and had been histopathologically found in patients after intraportal LCT [37]. Thus, MRI enabled noninvasive monitoring of microthrombi formation in the liver, which could not previously be detected by conventional imaging modalities. The engraftment of single MPIO-labeled liver cells into liver plates could not be visualized by MRI as the detection threshold was approximately 10,000 MPIOlabeled cells under the imaging conditions applied in this preclinical model. Thus, this study demonstrated that under conditions of clinical abdominal MRI, single-cell detection is not possible in the liver, but microthrombi formation could be sufficiently detected using MPIO-labeled cells. This finding contrasts with the results from small animal models, emphasizing the need for large animal models and careful interpretation of the results of experimental models for MRI monitoring of hepatocyte transplantation.
Figure 3: MRI monitoring of liver cell transplantation under preclinical
conditions. (A–C) 3.0 Tesla MRI following liver cell transplantation in swines.
(A) Intrasplenic injected micron-sized iron oxide-labeled hepatocytes induce distinct
areas of hypointensity in the spleen (arrows; 7 days after transplantation). (B) After
intraportal infusion, signal voids are visible in the liver, which are caused by
microthrombi of micron-sized iron oxide-labeled donor cells. (C) Administration of
native cells induced no signal voids (day 7 after application). (D–F) MRI-controlled
intrasplenic liver cell infusion in swines (3.0 Tesla MRI). (D) The arteriogram shows
the identification of the splenic artery for catheter-based cell infusion. MRI (E) prior
and (F) after cell administration shows a clear signal decrease of the spleen caused
by micron-sized iron oxide-labeled liver cells.
Please refer to [83] for further details on (A–C).
Please refer to [84] for further details on (D–F).
In a second study, Raschzok et al. investigated the feasibility of fast dynamic ‘real-time’ MRI monitoring during LCT to the spleen [84]. The spleen is the most common ectopic implantation site for hepatocytes. Furthermore, growing evidence suggests that hepatocytes translocate from the spleen to the liver, making the spleen attractive as a reservoir for transplanted hepatocytes. MPIOlabeled hepatocytes were infused into the spleen of swines via a catheter placed in the lineal artery (Figure 3D), and imaging was performed immediately prior to and during cell infusion. Images from static 3.0 Tesla MRI demonstrated significantly lower signal intensities and signal-tonoise ratios after cell infusion compared with the pretransplant images (Figure 3E & F). T2-weighted fast dynamic MRI enabled visualization of the signal decrease in the spleen during cell infusion, which was histologically correlated with the presence of labeled cells in splenic microvessels.
This study highlights the feasibility of real-time MRI monitoring during LCT to the spleen. Following optimization of the imaging protocols, this method may potentially be used to monitor hepatocyte infusion to the liver through the portal vein.
Challenges with MRI monitoring of LCT
Several limitations arise for iron oxide particle-based MRI monitoring of LCT. First, discrimination between particle-labeled transplanted cells and cells of the host, which have taken up free particles, is challenging. While differences due to the different particle load of transplanted cells and cells of the recipient could be visualized by high resolution MRI in the study from Shaprio et al. [80], studies using lower field MRI have not achieved comparable results [81]. Moreover, due to the same reasons it is unlikely that it will be possible to differentiate between iron particles in different cells in the liver using MRI.
Although MRI provides the best quality for cellular and tissue imaging, the lack of single-cell detection ability in the clinical set-up limits the use of this method. Moreover, clinical MRI is not able to differentiate particle-labeled living cells from labeled dead cells. MRI monitoring therefore seems more appropriate for the monitoring of cell clusters or microthrombi formation during cell infusion than for monitoring single-cell engraftment [83,84].
Considering the fact that the stability of the iron-based particles has not been proven in longterm studies, safety issues regarding the circulation and the metabolism of these particles generate further hurdles for the clinical use of this method. While the smaller SPIOs are degraded over time, MPIOs is thought to remain stable [69]. We assume that MIPOs could remain encapsulated in the recipient body. However, long-term animal studies are necessary to address this hypothesis.
Optical methods for cell tracking
Optical methods for imaging cell transplantation are generally based on bioluminescence or fluorescence signal detection [54]. Optical-based methods in cell tracking provide information regarding the cell-to-cell interactions and metabolism, both in vitro and in preclinical in vivo models, with high sensitivity [85]. Fluorescence labeling and bioluminescence imaging can be used for in vitro trafficking of the cells as long as there is an optical window for the targets. As the conventional intravital microscopes, with or without fluorescence imaging, have a tissue penetration capacity of only a few millimeters, the capabilities of in vivo cell tracking have been limited in rodents so far [86–88]. Using the near-infrared fluorescence spectrum, the signal penetration and detection can be increased to up to 2–3 cm due to reduced absorption by the blood (oxygenized and deoxygenized hemoglobin) and a marked reduction in the autofluorescence signal from the skin [89]. More recent developments in fluorescence imaging involve the use of quantitative fluorescence molecular tomography imaging, a modality that exclusively works in the 680–750 nm window of the near-infrared spectrum [90]. In this procedure, a fluorophore is injected into an anesthetized mouse, which subsequently is placed in the machines’ imaging chamber and scanned by a near-infrared pair of laser diodes that provide multiple source-detector projections. Laser excitation and fluorescence emission are detected by a thermoelectrically cooled charge-coupled device camera, and an automated data set analysis is then used. Since the total amount of probe in the region of interest can be automatically calculated, this technique has great potential for cell tracking and quantification of labeled cells in vivo [91].
As a first attempt at optical imaging in LCT, Koenig et al. demonstrated the repopulation of dipeptidylpeptidase IV+ (DPPIV+) hepatocytes in a DPPIV-deficient liver in rats following intrasplenic application. Using near-infrared fluorescence with a Cy5.5-conjugated antibody, liver repopulation by the transplanted liver cells could be visualized over a period of 16 weeks [92]. Although optical imaging can provide detailed information regarding the transplanted cells, the limited tissue penetration depth and limited 3D spatial resolution of this method currently hinder its translation to the clinic.
Conclusion & future perspective
At present, only radionuclide-based imaging has been clinically used for noninvasive monitoring of transplanted liver cells (Table 2). This modality enables the short-term analysis of the biodistribution of in vitro labeled cells and, by repetitive tracer application, the noninvasive visualization of the metabolic activity of hepatocytes transplanted to an ectopic implantation site. However, this technology has several limitations, the short radionuclide half-lives being presumably the strongest limiting factor in helping to understand the long-term fate of transplanted cells in patients. Future clinical studies will combine advantages of radionuclide imaging with tomographic image acquisition and, possibly, the combination with CT or MRI will optimize tracer localization. While optical-based imaging technologies are a valuable tool for experimental small animal studies on LCT, a noninvasive clinical application for these technologies is less likely and is mainly limited by the insufficient tissue penetration capacity. MRI-based tracking of iron oxide particle-labeled liver cells has demonstrated promising results in animal models and has already reached preclinical studies. While there are clear benefits of this imaging modality, such as the high soft tissue contrast and the absence of radiation, concerns remain regarding the long-term detectability of transplanted cells and the possible toxicity of iron oxide particles. Furthermore, it is evident that, particularly under conditions of clinical MRI, it is currently not possible to discriminate between labeled cells, alive or dead, and macrophages that take up free particles [93,94]. Moreover, single-cell detection has not yet been achieved under clinical conditions. Further studies will be necessary to evaluate the possible benefits of MR-based cell tracking in LCT. However, as MRI has already been clinically used for tracking SPIO-labeled islet cells [95], it is highly probable that this approach will also be clinically applicable for monitoring LCT. In view of the pros and cons of radionuclideand MR-based imaging, a combination of both approaches may achieve the optimal result [54]. A multimodal particle with an iron core and appropriate radionuclide labeling could enable sensitive detection of transplanted cells in the liver below the cutoff for MRI-based cell tracking. The iron particle would then enable the detection of microthrombi formation during cell infusions in clinical trials aiming to optimize cell engraftment, which cannot be sufficiently visualized by radionuclide-based imaging due to image saturation. Alternatively, particles with stronger effects on the magnetic field or MR protocols may be developed that enable the detection of single cells in the human body.
The clearance of hepatocytes following transplantation is a general limitation to all imaging techniques, as the contrast agent will be released from dying hepatocytes and can be taken up by other cells of the liver. The immunological interaction between the donor cells and the host is another issue that has not yet been addressed by imaging technology. While it has been assumed that cell transplantation would require more liberal immunosuppression regimes compared with whole-organ transplantation, the ideal cell tracking imaging modality should both be able to visualize the transplanted cells and define the immune status of the recipient. Although the ideal solution for clinical noninvasive monitoring of LCT has not yet been found, we can hope that imaging technology will help optimize liver cell administration and the clinical outcome of LCT.
Financial & competing interests disclosure
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
The institutional research budget, Forschungsförderung, was used for funding the writing assistance from American Journal Experts.

References
- Dhawan A, Puppi J, Hughes RD, Mitry RR. Human hepatocyte transplantation: current experience and future challenges. Nat. Rev. Gastroenterol. Hepatol. 7(5), 288–298 (2010).
- Dhawan A, Strom SC, Sokal E, Fox IJ. Human hepatocyte transplantation. Methods Mol. Biol. 640, 525–534 (2010).
- Hughes RD, Mitry RR, Dhawan A. Current status of hepatocyte transplantation. Transplantation 93(4), 342–347 (2012).
- Darwish AA, Sokal E, Stephenne X, Najimi M, de Goyet Jde V, Reding R. Permanent access to the portal system for cellular transplantation using an implantable port device. Liver Transpl. 10(9), 1213–1215 (2004).
- Fox IJ, Chowdhury JR, Kaufman SS et al. Treatment of the Crigler–Najjar syndrome Type I with hepatocyte transplantation. N. Engl. J. Med. 338(20), 1422–1426 (1998).
- Ambrosino G, Varotto S, Strom SC et al. Isolated hepatocyte transplantation for Crigler–Najjar syndrome Type 1. Cell Transplant. 14(2–3), 151–157 (2005).
- Dhawan A, Mitry RR, Hughes RD. Hepatocyte transplantation for liver-based metabolic disorders. J. Inherit. Metab. Dis. 29(2–3), 431–435 (2006).
- Allen KJ, Mifsud NA, Williamson R, Bertolino P, Hardikar W. Cell-mediated rejection results in allograft loss after liver cell transplantation. Liver Transpl. 14(5), 688–694 (2008).
- Lysy PA, Najimi M, Stephenne X, Bourgois A, Smets F, Sokal EM. Liver cell transplantation for Crigler–Najjar syndrome Type I: update and perspectives. World J. Gastroenterol. 14(22), 3464–3470 (2008).
- Grossman M, Rader DJ, Muller DW et al. A pilot study of ex vivo gene therapy for homozygous familial hypercholesterolaemia. Nat. Med. 1(11), 1148–1154 (1995).
- Dhawan A, Mitry RR, Hughes RD et al. Hepatocyte transplantation for inherited factor VII deficiency. Transplantation 78(12), 1812–1814 (2004).
- Muraca M, Gerunda G, Neri D et al. Hepatocyte transplantation as a treatment for glycogen storage disease Type 1A. Lancet 359(9303), 317–318 (2002).
- Lee KW, Lee JH, Shin SW et al. Hepatocyte transplantation for glycogen storage disease type IB. Cell Transplant. 16(6), 629–637 (2007).
- Sokal EM, Smets F, Bourgois A et al. Hepatocyte transplantation in a 4-year-old girl with peroxisomal biogenesis disease: technique, safety, and metabolic follow-up. Transplantation 76(4), 735–738 (2003).
- Beck BB, Habbig S, Dittrich K et al. Liver cell transplantation in severe infantile oxalosis – a potential bridging procedure to orthotopic liver transplantation? Nephrol. Dial. Transplant. 27(7), 2984–2989 (2012).
- Strom SC, Fisher RA, Rubinstein WS et al. Transplantation of human hepatocytes. Transplant. Proc. 29(4), 2103–2106 (1997).
- Horslen SP, McCowan TC, Goertzen TC et al. Isolated hepatocyte transplantation in an infant with a severe urea cycle disorder. Pediatrics 111(6 Pt 1), 1262–1267 (2003).
- Stéphenne X, Najimi M, Smets F, Reding R, de Ville de Goyet J, Sokal EM. Cryopreserved liver cell transplantation controls ornithine transcarbamylase deficient patient while awaiting liver transplantation. Am. J. Transplant. 5(8), 2058–2061 (2005).
- Puppi J, Tan N, Mitry RR et al. Hepatocyte transplantation followed by auxiliary liver transplantation – a novel treatment for ornithine transcarbamylase deficiency. Am. J. Transplant. 8(2), 452–457 (2008).
- Meyburg J, Das AM, Hoerster F et al. One liver for four children: first clinical series of liver cell transplantation for severe neonatal urea cycle defects. Transplantation 87(5), 636–641 (2009).
- Stéphenne X, Najimi M, Sibille C, Nassogne MC, Smets F, Sokal EM. Sustained engraftment and tissue enzyme activity after liver cell transplantation for argininosuccinate lyase deficiency. Gastroenterology 130(4), 1317–1323 (2006).
- Soriano HE, Wood RP, Kang D et al. Hepatocellular transplantation in children with fulminant liver failure. Hepatology 26, 239A (1997).
- Bilir BM, Guinette D, Karrer F et al. Hepatocyte transplantation in acute liver failure. Liver Transpl. 6(1), 32–40 (2000).
- Strom SC, Chowdhury JR, Fox IJ. Hepatocyte transplantation for the treatment of human disease. Semin. Liver Dis. 19(1), 39–48 (1999).
- Fisher RA, Strom SC. Human hepatocyte transplantation: worldwide results. Transplantation 82(4), 441–449 (2006).
- Habibullah CM, Syed IH, Qamar A, Taher-Uz Z. Human fetal hepatocyte transplantation in patients with fulminant hepatic failure. Transplantation 58(8), 951–952 (1994).
- Fisher RA, Bu D, Thompson M et al. Defining hepatocellular chimerism in a liver failure patient bridged with hepatocyte infusion. Transplantation 69(2), 303–307 (2000).
- Schneider A, Attaran M, Meier PN et al. Hepatocyte transplantation in an acute liver failure due to mushroom poisoning. Transplantation 82(8), 1115–1116 (2006).
- Khan AA, Habeeb A, Parveen N et al. Peritoneal transplantation of human fetal hepatocytes for the treatment of acute fatty liver of pregnancy: a case report. Trop. Gastroenterol. 25(3), 141–143 (2004).
- Baccarani U, Adani GL, Sanna A et al. Portal vein thrombosis after intraportal hepatocytes transplantation in a liver transplant recipient. Transpl. Int. 18(6), 750–754 (2005).
- Strom SC, Fisher RA, Thompson MT et al. Hepatocyte transplantation as a bridge to orthotopic liver transplantation in terminal liver failure. Transplantation 63(4), 559–569 (1997).
- Li ZR, Mao XH, Hu XX et al. Primary human hepatocyte transplantation in the therapy of hepatic failure: 2 cases report. Asian Pac. J. Trop. Med. 5(2), 165–168 (2012).
- Mito M, Kusano M, Kawaura Y. Hepatocyte transplantation in man. Transplant. Proc. 24(6), 3052–3053 (1992).
- Sterling RK, Fisher RA. Liver transplantation. Living donor, hepatocyte, and xenotransplantation. Clin. Liver Dis. 5(2), 431–460 (2001).
- Fitzpatrick E, Mitry RR, Dhawan A. Human hepatocyte transplantation: state of the art. J. Intern. Med. 266(4), 339–357 (2009).
- Soltys KA, Soto-Gutiérrez A, Nagaya M et al. Barriers to the successful treatment of liver disease by hepatocyte transplantation. J. Hepatol. 53(4), 769–774 (2010).
- Quaglia A, Lehec SC, Hughes RD et al. Liver after hepatocyte transplantation for liverbased metabolic disorders in children. Cell Transplant. 17(12), 1403–1414 (2008).
- Rajvanshi P, Kerr A, Bhargava KK, Burk RD, Gupta S. Studies of liver repopulation using the dipeptidyl peptidase IV-deficient rat and other rodent recipients: cell size and structure relationships regulate capacity for increased transplanted hepatocyte mass in the liver lobule. Hepatology 23(3), 482–496 (1996).
- Gustafson EK, Elgue G, Hughes RD et al. The instant blood-mediated inflammatory reaction characterized in hepatocyte transplantation. Transplantation 91(6), 632–638 (2011).
- Han B, Lu Y, Meng B, Qu B. Cellular loss after allogenic hepatocyte transplantation. Transplantation 87(1), 1–5 (2009).
- Touboul T, Hannan NR, Corbineau S et al. Generation of functional hepatocytes from human embryonic stem cells under chemically defined conditions that recapitulate liver development. Hepatology 51(5), 1754–1765 (2010).
- Espejel S, Roll GR, McLaughlin KJ et al. Induced pluripotent stem cell-derived hepatocytes have the functional and proliferative capabilities needed for liver regeneration in mice. J. Clin. Invest. 120(9), 3120–3126 (2010).
- Fisher RA, Mas VR. Cell transplant techniques: engraftment detection of cells. Methods Mol. Biol. 481, 97–105 (2009).
- Zanzonico P. Principles of nuclear medicine imaging: planar, SPECT, PET, multimodality, and autoradiography systems. Radiat. Res. 177(4), 349–364 (2012).
- Zanzonico P. Positron emission tomography: a review of basic principles, scanner design and performance, and current systems. Semin. Nucl. Med. 34(2), 87–111 (2004).
- Yoon JK, Park BN, Shim WY, Shin JY, Lee G, Ahn YH. In vivo tracking of 111In-labeled bone marrow mesenchymal stem cells in acute brain trauma model. Nucl. Med. Biol. 37(3), 381–388 (2010).
- Kircher MF, Gambhir SS, Grimm J. Noninvasive cell-tracking methods. Nat. Rev. Clin. Oncol. 8(11), 677–688 (2011).
- Gholamrezanezhad A, Mirpour S, Bagheri M et al. In vivo tracking of 111In-oxine labeled mesenchymal stem cells following infusion in patients with advanced cirrhosis. Nucl. Med. Biol. 38(7), 961–967 (2011).
- Nagata H, Nishitai R, Shirota C et al. Prolonged survival of porcine hepatocytes in cynomolgus monkeys. Gastroenterology 132(1), 321–329 (2007).
- Yang W, Mou T, Shao G, Wang F, Zhang X, Liu B. Copolymer-based hepatocyte asialoglycoprotein receptor targeting agent for SPECT. J. Nucl. Med. 52(6), 978–985 (2011).
- Chouhan M, Puppi J, Solanas E, Mitry RR, Dhawan A, Hughes RD. Hepatocyte labeling with 99mTc-GSA: a potential noninvasive technique for tracking cell transplantation. Int. J. Artif. Organs 35(6), 450–457 (2012).
- Bohnen NI, Charron M, Reyes J et al. Use of indium-111-labeled hepatocytes to determine the biodistribution of transplanted hepatocytes through portal vein infusion. Clin. Nucl. Med. 25(6), 447–450 (2000).
- Sauer R, Muller RG. Strahlenphysik. In: Radiologie. Kaufmann GW, Moser E, Sauer R (Eds). Elsevier, Urban & Fischer, Munich, Jena, Germany, 9–32 (2006).
- Srinivas M, Aarntzen EH, Bulte JW et al. Imaging of cellular therapies. Adv. Drug Deliv. Rev. 62(11), 1080–1093 (2010).
- Eich T, Eriksson O, Sundin A et al. Positron emission tomography: a real-time tool to quantify early islet engraftment in a preclinical large animal model. Transplantation 84(7), 893–898 (2007).
- Jiang H, Cheng Z, Tian M, Zhang H. In vivo imaging of embryonic stem cell therapy. Eur. J. Nucl. Med. Mol. Imaging 38(4), 774–784 (2011).
- Pearl J, Wu JC. Seeing is believing: tracking cells to determine the effects of cell transplantation. Semin. Thorac. Cardiovasc. Surg. 20(2), 102–109 (2008).
- Yaghoubi SS, Campbell DO, Radu CG, Czernin J. Positron emission tomography reporter genes and reporter probes: gene and cell therapy applications. Theranostics 2(4), 374–391 (2012).
- Puppi J, Modo M. Use of magnetic resonance imaging contrast agents to detect transplanted liver cells. Top. Magn. Reson. Imaging. 20(2), 113–120 (2009).
- Arbab AS, Janic B, Haller J, Pawelczyk E, Liu W, Frank JA. In vivo cellular imaging for translational medical research. Curr. Med. Imaging Rev. 5(1), 19–38 (2009).
- de Figueiredo EH, Borgonovi AF, Doring TM. Basic concepts of MR imaging, diffusion MR imaging, and diffusion tensor imaging. Magn. Reson. Imaging Clin. N. Am. 19(1), 1–22 (2011).
- Pagani E, Bizzi A, Di Salle F, De Stefano N, Filippi M. Basic concepts of advanced MRI techniques. Neurol. Sci. 29(Suppl. 3), 290–295 (2008).
- Medarova Z, Vallabhajosyula P, Tena A et al. In vivo imaging of autologous islet grafts in the liver and under the kidney capsule in non-human primates. Transplantation 87(11), 1659–1666 (2009).
- Weissleder R, Cheng HC, Bogdanova A, Bogdanov A Jr. Magnetically labeled cells can be detected by MR imaging. J. Magn. Reson. Imaging 7(1), 258–263 (1997).
- Modo M, Hoehn M, Bulte JW. Cellular MR imaging. Mol. Imaging 4(3), 143–164 (2005).
- Slotkin JR, Cahill KS, Tharin SA, Shapiro EM. Cellular magnetic resonance imaging: nanometer and micrometer size particles for noninvasive cell localization. Neurotherapeutics 4(3), 428–433 (2007).
- Modo M, Meade TJ, Mitry RR. Liver cell labelling with MRI contrast agents. Methods Mol. Biol. 481, 207–219 (2009).
- Puppi J, Mitry RR, Modo M, Dhawan A, Raja K, Hughes RD. Use of a clinically approved iron oxide MRI contrast agent to label human hepatocytes. Cell Transplant. 20(6), 963–975 (2011).
- Raschzok N, Muecke DA, Adonopoulou MK et al. In vitro evaluation of magnetic resonance imaging contrast agents for labeling human liver cells: implications for clinical translation. Mol. Imaging Biol. 13(4), 613–622 (2011).
- Arbab AS, Yocum GT, Kalish H et al. Efficient magnetic cell labeling with protamine sulfate complexed to ferumoxides for cellular MRI. Blood 104(4), 1217–1223 (2004).
- Morgul MH, Raschzok N, Schwartlander R et al. Tracking of primary human hepatocytes with clinical MRI: initial results with Tatpeptide modified superparamagnetic iron oxide particles. Int. J. Artif. Organs 31(3), 252–257 (2008).
- Luciani A, Parouchev A, Smirnov P et al. In vivo imaging of transplanted hepatocytes with a 1.5-T clinical MRI system – initial experience in mice. Eur. Radiol. 18(1), 59–69 (2008).
- Shapiro EM, Skrtic S, Koretsky AP. Sizing it up: cellular MRI using micron-sized iron oxide particles. Magn. Reson. Med. 53(2), 329–338 (2005).
- Raschzok N, Morgul MH, Pinkernelle J et al. Imaging of primary human hepatocytes performed with micron-sized iron oxide particles and clinical magnetic resonance tomography. J. Cell Mol. Med. 12(4), 1384–1394 (2008).
- Kammer NN, Billecke N, Morgul MH et al. Labeling of primary human hepatocytes with micron-sized iron oxide particles in suspension culture suitable for large-scale preparation. Artif. Organs. 35(4), E91–E100 (2011).
- Nkansah MK, Thakral D, Shapiro EM. Magnetic poly(lactide-co-glycolide) and cellulose particles for MRI-based cell tracking. Magn. Reson. Med. 65(6), 1776–1785 (2011).
- Raschzok N, Langer CM, Schmidt C et al. Functionalizable silica-based micron-sized iron oxide particles for cellular magnetic resonance imaging. Cell Transplant doi:10.3727/096368912X661382 (2013) (Epub ahead of print).
- Ju S, Teng GJ, Lu H et al. In vivo MR tracking of mesenchymal stem cells in rat liver after intrasplenic transplantation. Radiology 245(1), 206–215 (2007).
- Bos C, Delmas Y, Desmoulière A et al. In vivo MR imaging of intravascularly injected magnetically labeled mesenchymal stem cells in rat kidney and liver. Radiology 233(3), 781–789 (2004).
- Shapiro EM, Sharer K, Skrtic S, Koretsky AP. In vivo detection of single cells by MRI. Magn. Reson. Med. 55(2), 242–249 (2006).
- Leconte I, Pallu S, Abarca-Quinones J et al. MRI of iron-oxide labelled transplanted hepatocytes in mice: effect of treatment with cyclophosphamide. J. Magn. Reson. Imaging 32(2), 367–375 (2010).
- Shi XL, Gu JY, Han B, Xu HY, Fang L, Ding YT. Magnetically labeled mesenchymal stem cells after autologous transplantation into acutely injured liver. World J. Gastroenterol. 16(29), 3674–3679 (2010).
- Raschzok N, Teichgräber U, Billecke N et al. Monitoring of liver cell transplantation in a preclinical swine model using magnetic resonance imaging. Cell Med. 1(3), 123–135 (2010).
- Raschzok N, Pinkernelle J, Billecke N et al. Feasibility of fast dynamic MRI for noninvasive monitoring during ectopic liver cell transplantation to the spleen in a porcine model. AJR 198(6), 1417–1423 (2012).
- Perez VL, Caicedo A, Berman DM et al. The anterior chamber of the eye as a clinical transplantation site for the treatment of diabetes: a study in a baboon model of diabetes. Diabetologia 54(5), 1121–1126 (2011).
- Borot S, Crowe LA, Toso C, Vallée JP, Berney T. Noninvasive imaging techniques in islet transplantation. Curr. Diab. Rep. 11(5), 375–383 (2011).
- Cao YA, Bachmann MH, Beilhack A et al. Molecular imaging using labeled donor tissues reveals patterns of engraftment, rejection, and survival in transplantation. Transplantation 80(1), 134–139 (2005).
- Newton IG, Plaisted WC, Messina-Graham S et al. Optical imaging of progenitor cell homing to patient-derived tumors. Contrast Media Mol. Imaging 7(6), 525–536 (2012).
- Leevy WM, Gammon ST, Jiang H et al. Optical imaging of bacterial infection in living mice using a fluorescent near-infrared molecular probe. J. Am. Chem. Soc. 128(51), 16476–16477 (2006).
- Ntziachristos V, Tung CH, Bremer C, Weissleder R. Fluorescence molecular tomography resolves protease activity in vivo. Nat. Med. 8(7), 757–760 (2002).
- Ntziachristos V, Schellenberger EA, Ripoll J et al. Visualization of antitumor treatment by means of fluorescence molecular tomography with an annexin V-Cy5.5 conjugate. Proc. Natl Acad. Sci. USA 101(33), 12294–12299 (2004).
- Koenig S, Krause P, Hosseini AS et al. Noninvasive imaging of liver repopulation following hepatocyte transplantation. Cell Transplant. 18(1), 69–78 (2009).
- Winter EM, Hogers B, van der Graaf LM, Gittenberger-de Groot AC, Poelmann RE, van der Weerd L. Cell tracking using iron oxide fails to distinguish dead from living transplanted cells in the infarcted heart. Magn. Reson. Med. 63(3), 817–821 (2010).
- Rodriguez-Porcel M. In vivo imaging and monitoring of transplanted stem cells: clinical applications. Curr. Cardiol. Rep. 12(1), 51–58 (2010).
- Toso C, Vallee JP, Morel P et al. Clinical magnetic resonance imaging of pancreatic islet grafts after iron nanoparticle labeling. Am. J. Transplant. 8(3), 701–706 (2008).
• • Reviews the current state of clinical liver cell transplantation.
• Reports the first application of imaging technology and clinical liver cell transplantation.
• • Highlights the current state-of-the-art in the field of imaging cellular therapies.
• Contains a general protocol for labeling of liver cells with iron oxide particles.
• • Demonstrates the feasibility of in vivo imaging of transplanted liver cells by MRI.
• • Reports the first clinical application of MRI for monitoring of superparamagnetic iron oxide particle-labeled pancreatic islet cells transplanted to the liver.