Review Article - Imaging in Medicine (2013) Volume 5, Issue 3
Quantitative analysis of structural neuroimaging of mesial temporal lobe epilepsy
Negar Memarian1, Paul M Thompson1,2, Jerome Engel Jr1 and Richard J Staba*1
1Department of Neurology, Reed Neurological Research Center, Suite 2155, University of California, 710 Westwood Plaza, Los Angeles, CA 90095, USA
2Laboratory of Neuroimaging, University of California, Los Angeles, CA, USA
- *Corresponding Author:
- Richard J Staba
Department of Neurology, Reed Neurological Research Center
Suite 2155, University of California, 710 Westwood Plaza
Los Angeles, CA 90095, USA
Tel: +1 310 825 8479
Fax:+1 310 206 8461
E-mail: rstaba@mednet.ucla.edu
Abstract
Keywords
drug-resistant epilepsy; DTI; mesial temporal lobe epilepsy; morphometry; MRI; quantitative neuroimaging
Epilepsy is a chronic brain disorder characterized by an enduring predisposition to generate spontaneous epileptic seizures. Epilepsy affects nearly 3 million Americans, making it the third most common neurological disorder in the USA. Worldwide, an estimated 50 million individuals are affected by epilepsy, which accounts for 1% of the global burden of disease [1,2]. Between 60 and 80% of people with epilepsy will achieve seizure control with antiseizure drugs [3–6]. However, these statistics imply that as many as 40% of patients with epilepsy have seizures that are not adequately controlled by antiseizure drugs. The International League Against Epilepsy has proposed that drug-resistant epilepsy is a failure of two tolerated, appropriately chosen antiseizure drug trials to achieve sustained seizure freedom [7]. Research indicates that only a small percentage (<10%) of individuals with epilepsy benefit from subsequent drug trials after failing the first two [8,9]. It is not known why some seizures are or become resistant to medication, but several features are frequently associated with pharmacoresistant seizures [10–13]. Among them, the most common pathology and one commonly associated with pharmacoresistant limbic seizures is mesial temporal or hippocampal sclerosis (HS) [14,15]. The classic pattern of HS described by Bratz is associated with significant neuron loss and gliosis in subfield CA1 and prosubiculum [16], as well as the area between blades of dentate gyrus or ‘end folium’ [17]. There is less damage to dentate gyrus granule cells, CA3 and particularly CA2 pyramidal cells, and relative preservation of cells in subicular and parahippocampal gyrus. Patients with unilateral HS, which can be detected using MRI epilepsy protocols, have seizures that arise from or involve the affected mesial temporal lobe (MTL) structures that correspond to characteristic clinical signs and symptoms [18,19]. HS is often associated with widespread bilateral limbic and neocortical disturbances [20]. It may also be present with other lesions including, but not limited to, heterotopia of the temporal lobe, cortical dysplasia, cavernous angioma, tumor, contusion and cerebral infarctions [21,22]. The presence of these features, family history of epilepsy (genetic) and prolonged febrile seizures in infancy are consistent with MTL epilepsy (MTLE) with HS.
Accurate diagnosis is critical as drug-resistant epilepsies, such as MTLE with HS, and those with well-circumscribed epileptogenic lesions, known pathophysiology and predictable natural history can be treated successfully with surgery [8]. Successful surgical outcome – that is, seizure freedom or significant reduction of disabling seizures – depends on accurately delineating the epileptogenic zone that theoretically represents the brain areas necessary and sufficient for generating spontaneous seizures. The epileptogenic zone cannot be measured directly, but is inferred from presurgical diagnostic tests, such as video-EEG monitoring using scalp electrodes or, in some cases, intracranial grid or depth electrodes and neuroimaging. MRI is the preferred imaging modality to identify structural abnormalities responsible for the generation of spontaneous seizures confirmed by electrophysiological studies – that is, epileptogenic lesions. Conventional MRI does not reliably capture epileptogenic abnormalities such as HS or many types of malformations of cortical development; however, detection improves dramatically with an epilepsy MRI protocol and neuroradiologists who are knowledgeable about structural lesions that cause epilepsy [23]. In addition, quantitative analysis of MRI can aid in the detection of structural lesions and, since MRI plays such an important role in the diagnosis and management of epilepsy, some have proposed that MRI should be included in the classification of epilepsy etiologies [24].
Technical advances in neuroimaging have spurred the development of sophisticated analysis techniques. These techniques have been used chiefly in the basic research of epilepsy and have provided new information on structural abnormalities associated with drug-resistant epilepsy and MTLE with HS in particular. Evidence from structural MRI, along with electrophysiological and histological data and postsurgical seizure freedom, indicate that there is not one type of HS, and the ‘hippocampal-centric’ view of MTLE is being revised with greater emphasis on networks that include mesial temporal and extratemporal limbic structures. The purpose of this article is to review several MRI analysis techniques that have been used to quantify the spatial distribution and extent of structural brain abnormalities. The authors discuss the basis of voxel-based morphometry (VBM), surface-based morphometry (SBM) and pattern- based morphometry as well as cortical pattern matching (CPM), and the advantages and disadvantages of each. The focus is on the application of these techniques and detection of MRI structural abnormalities in patient studies of MTLE. Finally, the authors comment on the future of MRI morphometry and implications for basic research and clinical studies of drugresistant epilepsy.
Review of the literature
Quantitative structural imaging of epilepsy
The detection of pathological structural substrates that cause epilepsy has increased significantly with the advent of noninvasive, high-resolution MRI [22]. According to the International League Against Epilepsy Neuroimaging Commission, the essential indications for MRI include partial or secondarily generalized seizures and generalized seizures that do not remit with antiseizure drug treatment, and development of progressive neurological or neuropsychological deficits [25]. Furthermore, the Neuroimaging Commission recommends that imaging of epilepsy should include T1- and T2-weighted sequences to cover the whole brain in a minimum of two orthogonal planes, with a slice thickness of 1.5 mm or less to allow reformatting in any orientation and 3D reconstruction [25]. In patients with HS as the suspected epileptogenic lesion, images are acquired in an oblique coronal plane, perpendicular to the anterior–posterior axis of the hippocampus. The presence of HS on T1-weighted imaging generally appears as shrunken hippocampal gray matter (GM) and often loss of internal architecture ipsilateral to the site of seizure onset. In some cases, GM loss occurs bilaterally, but more extensive alterations are often found in the ipsilateral rather than the contralateral hippocampus. Assisting in the detection of HS are other MRI scans, such as T2-weighted and fluid-attenuated inversion recovery (FLAIR) sequences, which voids the water signal from ventricular and subarachnoid cerebral spinal fluid (CSF) and makes it easier to see signal changes associated with pathology in periventricular GM, such as in the hippocampus. Studies have found patients with histologically verified HS or neocortical lesions identified on conventional MRI correspond to increased signal intensity on T2-weighted and FLAIR sequences [26,27]. These studies and others that correlated MRI measures (e.g., T2 relaxation times) with neuronal and glial cell density suggest that increased signal intensity reflects increased interstitial fluid due to neuron loss and possibly gliosis [28–30]. Recent work, however, has not found a correlation between T2 relaxation time or normalized FLAIR intensity and glial immunostaining [30,31].
Advanced MRI analysis techniques that are described in the sections that follow can quantify the extent of GM loss, which is necessary in cases when HS asymmetry is subtle, and can also display the surface contours and spatial patterns of GM loss in hippocampus and other brain structures. Based on imaging studies of MTLE, we now know that GM loss extends beyond the affected hippocampus and includes cortical GM and white matter (WM) alterations in ipsilateral and contralateral temporal lobe and often extratemporal lobe structures.
Voxel-based morphometry techniques
In VBM, whole-brain digital structural images are compared on the basis of properties corresponding with groups of voxels. Voxel-based methods have been implemented in many different ways (e.g., [32–35]), but the general premise is to establish voxel-for-voxel correspondence across subjects through nonlinear registration of multiple subjects’ brain images to a standard anatomical template. Once the images are aligned into the same coordinate space, one can perform voxel-wise statistical comparisons of GM and WM volume or concentration across different subject groups [36]. GM volume can be derived from the number of GM voxels in the brain and GM concentration is the amount of GM per unit of intracranial volume. The concentration at a voxel is the volume of the GM in a neighborhood around that voxel divided by the overall volume of the structures in the same region around that voxel [37]. The standard VBM process typically involves four steps: spatial normalization, tissue segmentation, spatial smoothing and statistical analysis [38]. Spatial normalization involves applying a nonlinear registration to each subject’s T1-weighted MR image, so that local areas (voxels corresponding to anatomical brain areas) stretch or compress with respect to each other to match a group template. The deformed image is then segmented into tissue classes (i.e., GM, WM and CSF) based on the intensity in the image as well as a predetermined probability that a particular type of tissue will be found at a given location [37]. The segmented image is then spatially smoothed. The intensity in each voxel of the smoothed image is a local weighted average of GM, WM or CSF from adjacent voxels, which is generally expressed as a GM, WM or CSF concentration [38]. The final step of VBM involves voxel-wise statistical analysis that commonly includes group comparisons or correlations with covariates of interest [33,38].
The initial implementation of VBM was modified to avoid problems that arose when data from multiple subjects were not accurately aligned into the same coordinate space. In some cases, group differences in tissue volumes were inferred when images were not fully aligned across subjects and groups [33]. This occurs because the parameters of the normalization only encode smoothed, low-frequency deformations that may not fully align GM and non-GM structures (e.g., ventricles). Optimized VBM methods often involve an additional step, which multiplies the spatially normalized GM concentration (or other tissue class) by its relative volume before and after spatial normalization. This is referred to as modulation and enables comparisons of voxel-wise GM volume differences between groups [33].
Voxel-based methods are computationally efficient as they work directly on the voxel grid. Furthermore, the smoothing step in VBM methods suppresses confounding variations due to the high variability of gyral anatomy between individuals [38]. Hence, VBM techniques are useful in studies that quantitatively evaluate the spatial distribution and extent of statistically significant GM loss (or gain) in one subject group with respect to another, for example, patients compared with age- and sex-matched controls [36,39]. VBM has been successfully used to characterize structural abnormalities in patients with Parkinson’s disease [40], Alzheimer’s disease [39], schizophrenia [41] and, of course, epilepsy [38].
Studies of MTLE using automated VBM analysis consistently find GM loss in the hippocampus ipsilateral to – and, to a lesser extent, contralateral to – the site of seizure onset in patients with MRI evidence of unilateral HS [42–45]. VBM-based GM loss has also been detected in the amygdala, entorhinal cortex and parahippocampal gyri [46,47]. A review of the studies cited above and others indicates that GM reductions extend beyond the sclerotic MTL ipsilateral to seizure onset and often include other limbic structures, such as the thalamus, cingulate gyrus and orbitofrontal cortex, as well as areas of temporal and parietal neocortex, striatum and cerebellum [38]. Comparable results have been found using standard or optimized VBM methods, and optimized VBM may better detect subtle structural abnormalities [46]. Some studies observed that GM loss was greater in patients with MRI evidence of HS than those without [48,49], and VBM was effective in detecting GM alterations associated with cortical dysplasia that can accompany HS [50]. In other studies, GM reductions were more obvious in patients with left compared with right MTLE [51–53], although one study detected more prominent GM loss in patients with right-sided MTLE [50]. In addition to GM loss, several studies found WM loss adjacent to MTL structures ipsilateral to seizure onset and in some cases in the contralateral temporal lobe, as well as bilateral loss within the frontal lobe [43,45,54–56]. An analysis of GM volume and postsurgical seizure freedom in a cohort of patients with left MTLE and histological evidence for HS found patients who were not seizure free had reduced GM volumes in ipsilateral posterior MTL and the contralateral hippocampus compared with seizure-free patients [57].
Surface-based morphometry techniques
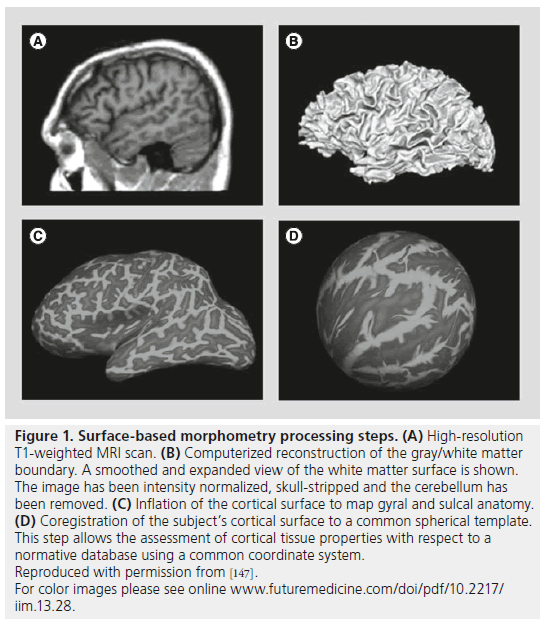
In SBM, morphometric measures are derived from geometric models of the cortical surface [37] or surface-based models of subcortical structures, such as the hippocampus. SBM techniques are used to reconstruct and analyze surfaces that represent structural boundaries within the brain. The major steps involved in SBM of the brain are preprocessing (image registration, intensity normalization, smoothing, brain extraction and brain segmentation), surface extraction, surface inflation or parameterization and surface mapping [58]. In order to align individual brain images and allow for comparisons across subjects, in cortical surface analyses, the brains are mapped to a unit sphere on which their original properties (e.g., cortical thickness) can be compared with each other, and results are mapped back to a reference brain surface (Figure 1). Several implementations of SBM have been made (e.g., [58–62]) with the FreeSurfer software from Harvard Martinos Center for Biomedical Imaging (MA, USA) being the most widely used (available through [201]). FreeSurfer is a set of automated tools for the reconstruction of the brain’s cortical surface from structural MRI data. It also allows the overlay of functional MRI data onto the reconstructed surface.
Figure 1. Surface-based morphometry processing steps. (A) High-resolution T1-weighted MRI scan. (B) Computerized reconstruction of the gray/white matter boundary. A smoothed and expanded view of the white matter surface is shown. The image has been intensity normalized, skull-stripped and the cerebellum has been removed. (C) Inflation of the cortical surface to map gyral and sulcal anatomy. (D) Coregistration of the subject’s cortical surface to a common spherical template. This step allows the assessment of cortical tissue properties with respect to a normative database using a common coordinate system. Reproduced with permission from [147]. For color images please see online www.futuremedicine.com/doi/pdf/10.2217/ iim.13.28.
Studies of MTLE using custom [63,64] and commercial SBM-based algorithms, such as FreeSurfer, have found significant hippocampal GM volume loss ipsilateral and, in some cases, contralateral to seizure onset [65,66]. However, in other studies, automated SBM analysis underestimated the extent of hippocampal GM loss compared with manual segmentation methods [67,68]. Patient studies of MTLE with HS found reduced ipsilateral hippocampal GM volumes correlated with cortical GM thinning in the temporal lobe [66,69]. Cortical GM was on average 5–15% thinner in bilateral frontal (prominently within pre- and para-central gyri operculum) and temporal lobes (Heschl’s gyrus) of patients with MTLE compared with controls [70], with somewhat greater thinning in patients with left- versus right-sided MTLE [71]. Furthermore, reduced thalamic GM volume extending from anterior to posterior primarily along the medial surface correlated with reduced hippocampal volume (subfield CA1) and MTL GM thinning [72], as well as bilateral frontal–central and lateral temporal lobe areas [73]. One study evaluated the presurgical presence of MRI hippocampal atrophy and postsurgical seizure freedom and found that, in cases with hippocampal atrophy, patients with postsurgical seizures had ipsilateral temporopolar and bilateral insular atrophy compared with patients who were seizure free [74]. Interestingly, in cases without hippocampal atrophy, postsurgical seizures were associated with ipsilateral posterior lateral temporal and contralateral parietal–occipital lobe atrophy.
Other cortical morphometry techniques
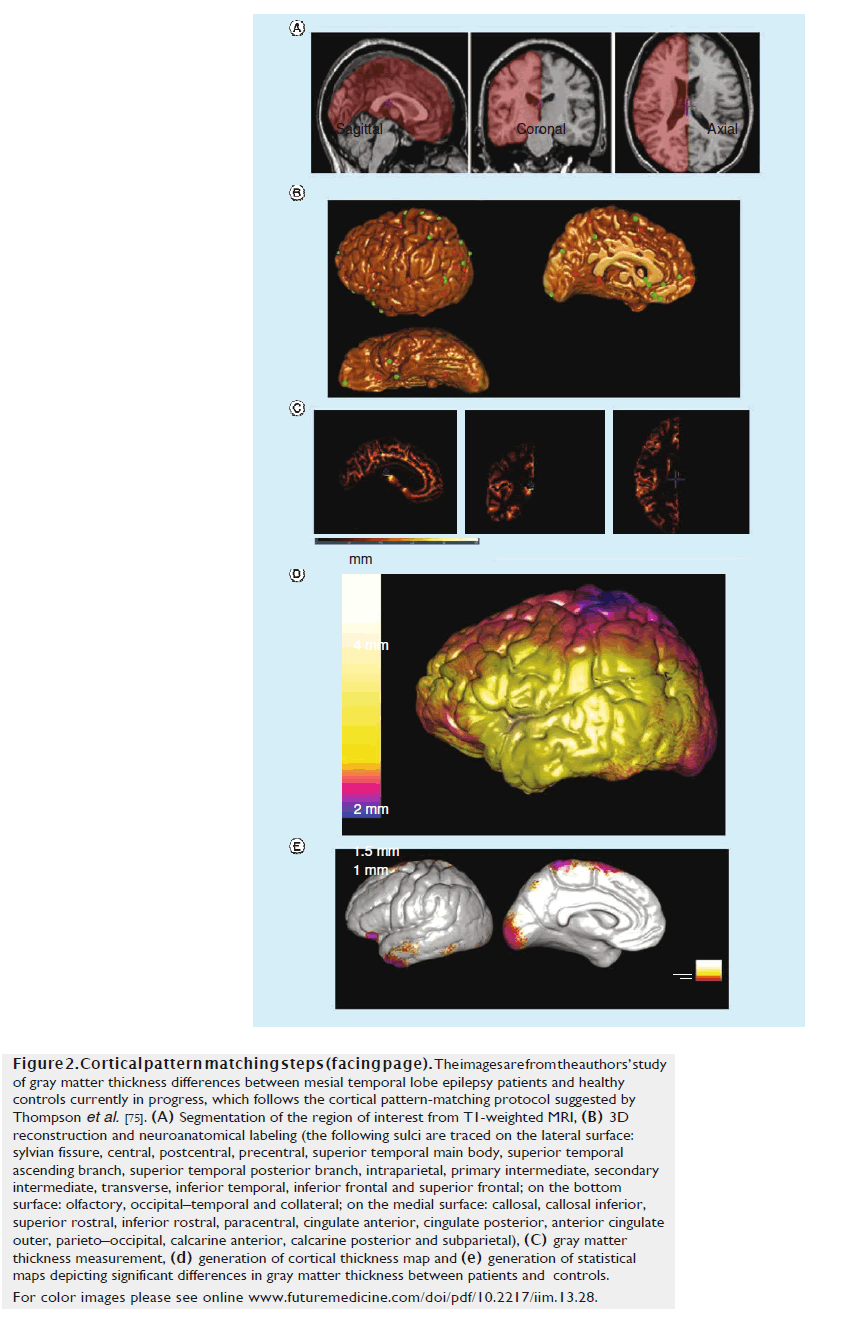
CPM is related to SBM, but differs in that after creating explicit geometric models of the cortex using parametric surfaces, deformation maps are built on the geometric models to spatially align cortical regions across subjects [75,76]. This is done by applying mathematical transformations that align key anatomical landmarks from one dataset to another. CPM techniques usually involve three main steps of cortical parameterization, matching cortical features across individuals and statistical comparisons with map group differences in features such as cortical thickness or GM concentrations. The first step involves creating geometrical models of the cortical surface and includes several steps, such as image segmentation, registration and 3D reconstruction. Matching cortical features across individuals is performed through neuroanatomical labeling (e.g., marking sulcal lines) and warping one brain surface onto another. Finally, statistical testing is performed to investigate the effect of various factors (e.g., disease, aging, treatment and genetics, among others) on various cortical attributes, such as GM thickness or gyrification [77]. These steps are shown in Figure 2 from a study of GM thickness differences between MTLE patients and healthy controls.
Figure 2. Cortical pattern matching steps (facing page). The images are from the authors’ study of gray matter thickness differences between mesial temporal lobe epilepsy patients and healthy controls currently in progress, which follows the cortical pattern-matching protocol suggested by Thompson et al. [75]. (A) Segmentation of the region of interest from T1-weighted MRI, (B) 3D reconstruction and neuroanatomical labeling (the following sulci are traced on the lateral surface: sylvian fissure, central, postcentral, precentral, superior temporal main body, superior temporal ascending branch, superior temporal posterior branch, intraparietal, primary intermediate, secondary intermediate, transverse, inferior temporal, inferior frontal and superior frontal; on the bottom surface: olfactory, occipital–temporal and collateral; on the medial surface: callosal, callosal inferior, superior rostral, inferior rostral, paracentral, cingulate anterior, cingulate posterior, anterior cingulate outer, parieto–occipital, calcarine anterior, calcarine posterior and subparietal), (C) gray matter thickness measurement, (D) generation of cortical thickness map and (E) generation of statistical maps depicting significant differences in gray matter thickness between patients and controls. For color images please see online www.futuremedicine.com/doi/pdf/10.2217/iim.13.28.
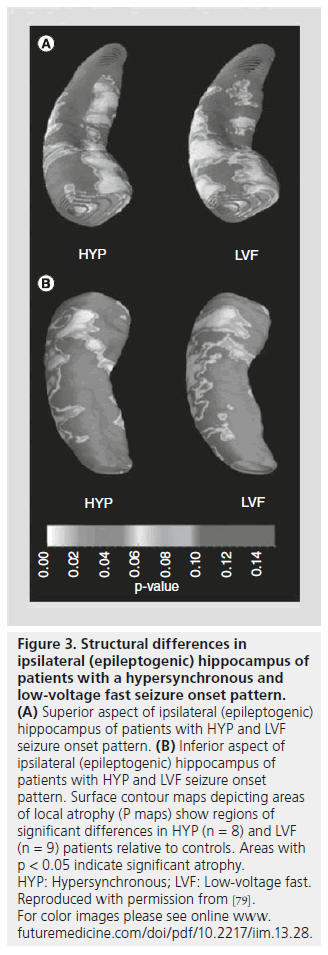
Similar to the studies using VBM and SBM methods described in the preceding sections, CPM techniques have been used to evaluate neocortical GM thickness in patients with MTLE and HS. In one study, GM thickness was reduced by up to 30% and reductions in cortical complexity were found in patients compared with controls [20]. In this latter study, significant GM loss was observed bilaterally in frontal areas (pole, operculum and orbitofrontal areas), lateral temporal and occipital regions independent of the hemisphere of seizure onset. In patients with HS, analyses of hippocampal surface structure found significant alterations on the lateral hippocampal surface corresponding to Sommer’s sector, while patients without HS had surface changes on the superior surface of the hippocampal body, in right-side MTLE, and on the superior surface of the medial hippocampal head region, near the uncinate gyrus, in left-sided MTLE [78]. Another study evaluated the spatial distribution of hippocampal atrophy in relation to different depth electroderecorded ictal EEG onset patterns [79]. In this study, based on EEG morphology, propensity for seizure spread and ipsilateral histological features, it was hypothesized that seizures beginning with hypersynchronous and low-voltage fast EEG patterns reflect different neuronal and anatomical mechanisms of generation. Results of CPM analysis found that patients with hypersynchronous ictal onsets had significant hippocampal atrophy ipsilateral to seizure onset that resembled a classic pattern of HS (sparing of subfield CA2; Figure 3), while patients with lowvoltage fast ictal onset had diffuse ipsilateral hippocampal atrophy and significant atrophy in contralateral hippocampus. Interestingly, in a separate study, surgical patients with MTLE who were not seizure free had atrophy that was more diffuse in the ipsilateral hippocampus and significant atrophy in anterior and lateral areas of the contralateral hippocampus compared with patients who were seizure free [80].
Figure 3.Structural differences in ipsilateral (epileptogenic) hippocampus of patients with a hypersynchronous and low-voltage fast seizure onset pattern. (A) Superior aspect of ipsilateral (epileptogenic) hippocampus of patients with HYP and LVF seizure onset pattern. (B) Inferior aspect of ipsilateral (epileptogenic) hippocampus of patients with HYP and LVF seizure onset pattern. Surface contour maps depicting areas of local atrophy (P maps) show regions of significant differences in HYP (n = 8) and LVF (n = 9) patients relative to controls. Areas with p < 0.05 indicate significant atrophy. HYP: Hypersynchronous; LVF: Low-voltage fast. Reproduced with permission from [79]. For color images please see online www. futuremedicine.com/doi/pdf/10.2217/iim.13.28.
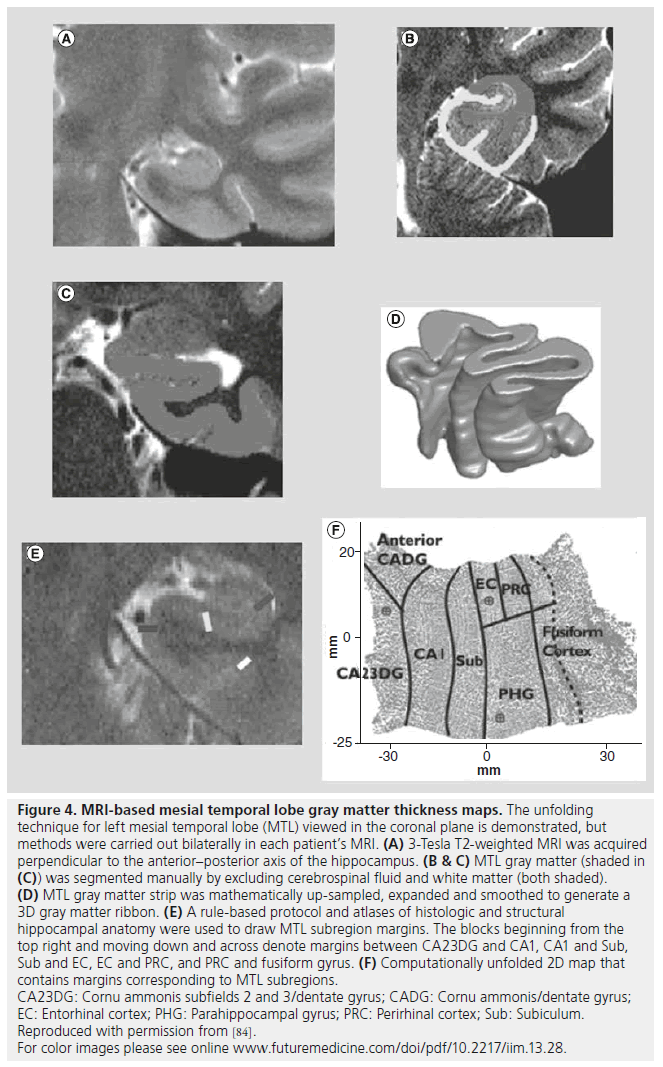
Due to the complex folding of the human MTL, it is difficult to visualize the hippocampal formation (dentate gyrus, hippocampus subfields CA1-3 and subicular cortex) together with adjacent structures (entorhinal cortex, parahippocampal gyrus and fusiform gyrus). Advances have been made with a novel cortical unfolding technique using high-resolution T2-weighted MRI that increases signal-to-noise ratio and resolution of MTL structures [81,82]. Figure 4 illustrates the main steps in the unfolding procedure that includes the standard manual segmentation of GM, WM and CSF, and an interpolation step to generate a 3D GM ribbon corresponding to MTL structures. The 3D GM ribbon is then computationally unfolded and flattened into 2D and structural boundaries projected onto the flattened image to produce an anatomical map of the adjoining MTL subregions. Similar to other morphometry techniques, GM thickness can be computed for each of the MTL subregions and individual maps of the MTL warped into a common plane with maps from other subjects to compute group averaged GM thickness maps that retain much of the anatomical variability within the subject group. In addition, individual maps provide the capability to localize and characterize functional MRI signal changes simultaneously across MTL subregions [81,83]. Furthermore, MTL maps generated from MRI of presurgical patients can be used to locate the position of multiple microelectrodes extending beyond the distal tip of clinical depth electrodes (Figure 4F; red circles depict location of microelectrodes) [82]. One study applied these techniques to evaluate interictal MTL single neuron firing properties in relation to subregion GM loss computed in patients with respect to controls [84]. Results from this study found significant bilateral MTL GM loss, although, compared with MTL contralateral to seizure onset, there was a stronger correlation between GM loss and increased neuronal firing in the ipsilateral MTL. Adjusting for levels of GM loss revealed significantly higher neuronal firing and bursting in ipsilateral than contralateral MTL, suggesting that synaptic reorganization following cell loss is associated with varying degrees of epileptogenicity.
Figure 4.MRI-based mesial temporal lobe gray matter thickness maps. The unfolding technique for left mesial temporal lobe (MTL) viewed in the coronal plane is demonstrated, but methods were carried out bilaterally in each patient’s MRI. (A) 3-Tesla T2-weighted MRI was acquired perpendicular to the anterior–posterior axis of the hippocampus. (B & C) MTL gray matter (shaded in (C)) was segmented manually by excluding cerebrospinal fluid and white matter (both shaded). (D) MTL gray matter strip was mathematically up-sampled, expanded and smoothed to generate a 3D gray matter ribbon. (E) A rule-based protocol and atlases of histologic and structural hippocampal anatomy were used to draw MTL subregion margins. The blocks beginning from the top right and moving down and across denote margins between CA23DG and CA1, CA1 and Sub, Sub and EC, EC and PRC, and PRC and fusiform gyrus. (F) Computationally unfolded 2D map that contains margins corresponding to MTL subregions. CA23DG: Cornu ammonis subfields 2 and 3/dentate gyrus; CADG: Cornu ammonis/dentate gyrus; EC: Entorhinal cortex; PHG: Parahippocampal gyrus; PRC: Perirhinal cortex; Sub: Subiculum. Reproduced with permission from [84]. For color images please see online www.futuremedicine.com/doi/pdf/10.2217/iim.13.28.
Pattern-based techniques
A more recent approach to study brain structure involves data-driven techniques that use machine learning to extract specific patterns [85], features [86] or objects [87] in MRI. Unlike most morphometry methods that assume one-to-one correspondence of anatomy across subjects, pattern-based methods identify distinctive anatomical patterns (e.g., appearance and geometry) that could be due to disease and, thus, are only present in subsets of subjects. These features can be automatically extracted from a set of subject images and their relationship to diagnostic categories can be modeled using supervised learning. Pattern-based techniques seek to identify instances of the same image feature in different subjects and discover features that tend to co-occur with specific subject groups [86]. Pattern-based techniques have been clinically validated for identification of structural differences between normal and Alzheimer’s disease patients [85,86]. They have also been used for gyrus-based parcellation of the cortical surface [87,88]. However, the pattern-based approaches have not yet been used in epilepsy studies.
Alternative MRI modalities
Lesions may not be obvious on conventional T1-weighted MRI in as many as 20% of patients with pharmacoresistant seizures [89], although alternative techniques, such as T2-weighted MRI, FLAIR and other MRI sequences, including double inversion recovery [90,91] and magnetization transfer ratio [90], can detect structural abnormalities in approximately half of these patients [92]. In addition, a special type of MRI is sensitive to the diffusion of water using pulsed field gradients with a method called diffusionweighted imaging. The extent of water movement freedom is quantified in a parameter called apparent diffusion coefficient (ADC). Several retrospective diffusion-weighted imaging studies found increased ADC values in the temporal lobe including the hippocampus and temporal pole [93–96] and posterior aspects of corpus callosum [97]. In many studies, ADC values were appropriately lateralized to the site of seizure onset in a large percentage of patients with MTLE and HS, although measures of ADC alone and some of the MRI methods described above had lower specificity for the seizure onset zone in patients without HS [98–100].
A special type of diffusion-weighted imaging is diffusion tensor imaging (DTI), which provides several useful measures of the restricted movement of water in tissue. Parameters derived from DTI include mean diffusivity, which reflects the overall rate of water diffusion in a voxel, and fractional anisotropy (FA), which measures how directionally constrained the diffusion process is, and is an indirect measure of WM (fiber) density or myelination levels. By following the principal directions of water diffusion, tract tracing methods can be used to follow neural pathways throughout the brain, and infer axonal connections. DTI studies have mapped the patterns of connectivity between the parahippocampal gyrus and orbitofrontal areas, as well as direct connectivity between the parahippocampal gyrus and the hippocampus itself (Figure 5A) [101]. The connections between neocortical areas and the hippocampus via the parahippocampal gyrus may provide a structural basis for theoretical models of seizure propagation. In patients with MTLE, voxel-based analysis of DTI has found increased mean diffusivity in the ipsilateral hippocampus, parahippocampal gyrus and thalamus [102–104]. Reduced FA was observed in ipsilateral temporal lobe including hippocampus and parahippocampal gyrus, areas of frontal lobe and portions of corpus callosum [105–107]. In several studies, reductions in FA were more prominent ipsilateral and, in some cases, contralateral to seizure onset in left compared with right MTLE [105,108,109]. In patients without lesions on MRI, reduced FA was found primarily in ipsilateral temporal lobe, corpus callosum and thalamus [104,110]. DTI studies using tractography found reductions in fornix bilaterally and cingulum [111,112], as well as increased mean diffusivity in ipsilateral WM fibers of the uncinate, arcuate and inferior longitudinal fasciculi, with the latter changes correctly lateralized to the site of seizure onset in 91% of patients with MTLE and hippocampal atrophy [111]. DTI has also been utilized to determine the prospective functional deficit zone in MTLE surgery candidates [113,114].
Figure 5.Diffusion tensor imaging for imaging structural connectivity. (A) Averaged connectivity maps from the left parahippocampal gyrus in ten controls, superimposed on the normalized single T1 images from the MRI provided by statistical parametric mapping 99 (sagittal views). These maps are displayed without applying any threshold to the connectivity values and display connectivity between the parahippocampal gyrus and the anterior temporal lobe, orbitofrontal areas, posterior temporal lobe and extrastriate occipital lobe via the lingual and fusiform gyri. (B) Results from probabilistic tractography obtained from different regions of interest averaged over all subjects. Each 3D brain rendering illustrates the voxel-wise probabilistic map of connectivity from one region of interest (top left box: left anterior cingulate; top right box: left inferior orbital region; bottom left box: left hippocampus; and bottom right box: left thalamus). (A) Reproduced with permission from [92] and (B) reproduced with permission from [127]. For color images please see online www.futuremedicine.com/doi/pdf/10.2217/ iim.13.28.
Discussion
None of the structural imaging techniques reviewed here have been used in the clinical presurgical evaluation of epilepsy, although each of the imaging techniques mentioned has strengths and weaknesses that are currently suitable for different research applications. VBM is relatively simple to implement, free and widely used in MRI studies of MTLE. While VBM is generally accurate for measuring GM atrophy in structures below the pial surface (e.g., hippocampus [38,44,115,116], amygdala [117] and thalamus [38,48,118]) and in some studies cingulate gyrus [118,119], it does not consistently detect GM abnormalities in the neocortex. This is due to the wide variations in cortical patterning across subjects, which makes it challenging to automatically align anatomical landmarks and cortical regions across subjects. In addition, it is difficult to detect significant differences in GM associated with neocortical pathology that is subtle or spatially complex [120]. SBM, on the other hand, is well-suited for detecting neocortical GM abnormalities because images are aligned with respect to neocortical folding patterns, and this improves neocortical registration compared with aligning on the basis of voxel intensity [121]. SBM methods also have a somewhat higher reproducibility than VBM and can be more robust in longitudinal studies that measure changes in GM thickness over time [36]. CPM uses high dimensional registration to provide detailed measurements of GM thickness in the hippocampus or neocortex of patients with MTLE. CPM is ideal for capturing small differences in GM thickness in patients without obvious pathology on MRI, although measures of volumetric GM loss can be sensitive to the geometrical calibration of the MRI scanner [121]. Furthermore, CPM can be difficult to implement and CPM algorithms are not as readily available or easy to use as VBM or SBM algorithms. Based on the differences in the design and application between these analysis techniques, it is difficult to specify one as the gold standard and use it in validation studies of other analysis techniques. The most reliable means of validation for these methods are correlation studies that relate measures of GM thickness to histological measures of neuronal and glial cell density.
Results from imaging analysis studies reflect statistically consistent differences across a sample of patients with MTLE typically with respect to control subjects. However, it is not known how accurate group-averaged data correspond to the individual patient, which is the basis for clinical decisions. More studies are required to determine the implication of these findings in the surgical outcome of patients who undergo epilepsy surgery. So far, several studies have shown that VBM in particular may not be a suitable standalone technique for detecting or spatially characterizing focal lesions in individual patients [122–124]. Due to the low specificity of the findings, caution is especially needed when evaluating MRI-negative patients for localization of the epileptogenic zone [99]. In a VBM-based analysis of GM in patients with left MTLE and HS and other MRI-negative patients, GM abnormalities were detected in only 20% of patients with HS and 20% of MRInegative patients [124]. More recently, VBM analysis on single case studies that compared single subjects with a control group found a very high false-positive rate [123]. This can be because any significant difference may be driven by individual variability in neuroanatomy rather than the neuropathology of the disease under investigation, or may represent a false positive due to the data being sampled from non-normally distributed populations [123].
Supervised learning algorithms are a potential strategy to incorporate quantitative MRI techniques in surgical decision-making for individual patients. Supervised learning algorithms can be trained by a set of multimodal features (e.g., EEG, MRI and clinical information) for each patient and their corresponding surgery outcome result. These algorithms seek to find patterns between features that are exclusively associated with the given outcome. Usually, such patterns are not obvious from visual inspection of the data. After the algorithm learns from a group of patients, it can then classify a new patient (i.e., predict surgery outcome) based on his/her multimodal features. Examples of this approach in MTLE surgical decision-making can be found in [125] and [126]. One retrospective study that used support vector machine DTI classification achieved high diagnostic accuracy in lateralizing the seizure onset zone in individual subjects [115]. Other studies using novel computer-automated SBM and DTI algorithms have produced results comparable with manual, labor-intensive methods [63,68,108]. Ultimately, these strategies could be used during presurgical evaluation to help plan the placement of intracranial grid or depth electrodes to determine sites of seizure onset or provide information on fiber pathways prior to resective surgery to minimize the risk of cognitive, motor or visual-field impairment [92].
Considering the strengths and weaknesses of the MRI analysis techniques discussed in the preceding paragraphs, it appears that significant structural abnormalities extend beyond the epileptogenic hippocampus in patients with MTLE and HS. In addition, MRI studies have found factors such as hemisphere of seizure onset, duration of epilepsy, age of onset, history of febrile seizures and possibly extent of HS and comorbidity to explain some of the variability in spatial patterns of extra-hippocampal GM loss, as these could be important for identifying brain areas consistently damaged in patients with MTLE [42,59,79,80]. Recent work has used graph theoretical analysis to evaluate anatomical networks and results are consistent with the hypothesis that MTLE is a network disease of reorganized mesial temporal and extratemporal limbic structures (Figure 5B) [127,128]. What is not known is whether all or only some areas of the reorganized network reflect the epileptogenic zone. Remote GM loss found in many of the MRI studies could reflect, for example, deafferentation from the MTL where seizures begin [118], excitotoxic neuron loss from propagated seizure activity, developmental abnormalities that contribute to alterations in extra-hippocampal connectivity and structure [129], GM loss due to an initial injury and/or progression of the damage caused by the initial injury, or chronic exposure to antiseizure drugs [20]. Some of these structural alterations may be epileptogenic, while others may not. The structural analysis techniques mentioned here, such as cortical unfolding or hippocampal CPM, that can localize electrophysiological or functional MRI signals and relate it to measures of GM thickness could help distinguish epileptogenic from nonepileptogenic lesions [81,83,130].
Determining the boundaries of the epileptogenic zone has clear implications in the surgical treatment and prognosis for postsurgical seizure freedom. A review of epilepsy surgery found that the percentage of surgical patients with HS who were seizure free was between 48 and 84% [131]. This latter review and others cite several factors associated with seizure freedom [18], including subtypes of HS. A histological study by Blumcke and colleagues found that patients with less severe HS (e.g., end folium sclerosis) were more likely to have postsurgical seizures compared with patients with the classic pattern of HS who were more likely to be seizure free [132]. A technical explanation for these histological results could be that MRI does not reliably detect the full spatial distribution of damage associated with more subtle forms of HS. Results from a VBM study found residual lesions posterior to ipsilateral hippocampus and/or in the contralateral hippocampus in patients who received anterior temporal resection that were presumed to be the basis for persistent postsurgical seizures [57]. By contrast, a recent randomized trial for early resective surgery found that, in a carefully selected group of patients with unilateral MTLE, neuroimaging indicative of HS, and an absence of MRI and EEG evidence for remote epileptogenic lesions, the percentage of seizure-free patients was 85% at 2 years after surgery [133]. Although histology did not confirm the subtype of HS in this trial, the high percentage of postsurgical seizure freedom is consistent with a homogenous cohort of classical HS.
Conclusion
Basic research studies of drug-resistant epilepsy using semiautomated structural MRI techniques provide noninvasive, efficient and complementary approaches for identifying and quantifying different types of anatomical abnormalities. In cases of MTLE, evidence from structural MRI studies suggest that the brain areas responsible for generating spontaneous seizures include the sclerotic hippocampus, as well as other limbic brain areas. How well spatial patterns of GM loss derived from group-averaged data correspond to an individual subject and whether all GM abnormalities are epileptogenic is not known and complicates the translation of these techniques to clinical epilepsy. We anticipate that MRI and other brain imaging studies, such as DTI and functional MRI using connectivity analysis combined with electrophysiological recordings and clinical history of epilepsy and comorbidity, will provide new information on the spatial distribution of the epileptogenic zone corresponding to different types of MTLE with HS.
Future perspective
Improvements in image acquisition & processing
Improvements in image acquisition technology, primarily improving the spatial resolution and signal-to-noise ratio of images, will be a major area of future work. Enhancing the accuracy and computational speed of quantitative neuroimaging algorithms is also important. With an increasing number of studies using automated SBM, there is a clear need for more accurate segmentation algorithms, particularly for segmenting subcortical regions. Invention of advanced methods for efficient registration of multiple structural imaging modalities, as well as integrating structural and functional imaging data, are crucial for the development of multimodal imaging studies of epilepsy. These improvements will benefit epilepsy research and possibly preoperative and prognosis assessments, as well as image-guided depth electrode implantation and resection surgery.
Multimodal imaging studies of MTLE
Multimodal imaging of MTLE through integration of structural and functional imaging data is another important area of future work. Functional imaging modalities such as PET [134–137] and SPECT [138] can be useful in determining the prospective functional deficit zone [92]. Functional MRI [139–142] and magnetic source imaging or magnetoencephalography [143,144] can yield more information about the ictal onset zone and the spatial extent of interictal spiking, often termed the ‘irritative zone’ [8]. The combination of functional imaging modalities with structural neuroimaging and techniques that spatially coregister functional and structural signals (e.g., [81,82]) may elucidate the functional role of damaged hippocampal and extra-hippocampal brain areas in the progression of the disease and the surgical outcome of patients.
Constructing a network of the epileptogenic zone
The structural and functional connectivity corresponding to the epileptogenic network is not known. More advanced MRI studies, along with graph theory-based models, are required to shed light on the anatomical and functional connectivity of epileptogenic structures and reveal potential seizure propagation pathways. With studies finding extra-hippocampal neocortical atrophy, further imaging and electrophysiological investigations are crucial to verifying whether those areas reflect damage due to propagated seizure activity, or if they are morphological changes that independently support the generation of spontaneous seizures.
Emerging field of connectomics
The emerging science of connectomics provides an approach to understanding large-scale brain networks in which normal and abnormal brain functions operate [145,146]. Progress in this field might elucidate the yet unknown mechanisms of seizure onset and termination. Furthermore, the combination of connectomics with biologically inspired dynamic models of epilepsy provides a suitable platform to explore the dynamic network nature of epilepsy. This can eventually improve epilepsy surgery by identifying specific connections or network nodes that, if cut or resected, will abolish seizures.
Acknowledgements
The authors would like to thank A Reid for her valuable comments on this manuscript.

Financial & competing interests disclosure
This work was supported by the Natural Sciences and Engineering Research Council of Canada (N Memarian), and Grant NS071048 from the NIH (R Staba). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
* of interest
* of considerable interest
- Hirtz D, Thurman DJ, Gwinn-Hardy K, Mohamed M, Chaudhuri AR, Zalutsky R. How common are the ‘common’ neurologic disorders? Neurology 68(5), 326–337 (2007).
- Murray CJ, Lopez AD, Jamison DT. The global burden of disease in 1990: summary results, sensitivity analysis and future directions. Bull. World Health Organ. 72(3), 495–509 (1994).
- Devinsky O. Patients with refractory seizures. N. Engl. J. Med. 340(20), 1565–1570 (1999).
- Brodie MJ, Barry SJ, Bamagous GA, Norrie JD, Kwan P. Patterns of treatment response in newly diagnosed epilepsy. Neurology 78(20), 1548–1554 (2012).
- Kwan P, Brodie MJ. Drug treatment of epilepsy: when does it fail and how to optimize its use? CNS Spectr. 9(2), 110–119 (2004).
- Sander JW. Some aspects of prognosis in the epilepsies: a review. Epilepsia 34(6), 1007–1016 (1993).
- Kwan P, Arzimanoglou A, Berg AT et al. Definition of drug resistant epilepsy: consensus proposal by the ad hoc Task Force of the ILAE Commission on therapeutic strategies. Epilepsia 51(6), 1069–1077 (2010).
- Engel J Jr. Seizures and Epilepsy. Oxford University Press, NY, USA (2013).
- Kwan P, Sperling MR. Refractory seizures: try additional antiepileptic drugs (after two have failed) or go directly to early surgery evaluation? Epilepsia 50(Suppl. 8), S57–S62 (2009).
- Go C, Snead OC 3rd. Pharmacologically intractable epilepsy in children: diagnosis and preoperative evaluation. Neurosurg. Focus 25(3), E2 (2008).
- Berg AT, Shinnar S, Levy SR et al. Defining early seizure outcomes in pediatric epilepsy: the good, the bad and the in-between. Epilepsy Res. 43(1), 75–84 (2001).
- Stephen LJ, Kelly K, Mohanraj R, Brodie MJ. Pharmacological outcomes in older people with newly diagnosed epilepsy. Epilepsy Behav. 8(2), 434–437 (2006).
- Hitiris N, Mohanraj R, Norrie J, Sills GJ, Brodie MJ. Predictors of pharmacoresistant epilepsy. Epilepsy Res. 75(2–3), 192–196 (2007).
- Blumcke I, Thom M, Wiestler OD. Ammon’s horn sclerosis: a maldevelopmental disorder associated with temporal lobe epilepsy. Brain Pathol. 12(2), 199–211 (2002).
- Engel J Jr. Surgical treatment for epilepsy: too little, too late? JAMA 300(21), 2548–2550 (2008).
- Bratz E. [Ammon horn findings in epileptics]. Arch. Psychiatr. Nervenkr. 32, 820–835 (1899).
- Margerison JH, Corsellis JA. Epilepsy and the temporal lobes. A clinical, electroencephalographic and neuropathological study of the brain in epilepsy, with particular reference to the temporal lobes. Brain 89(3), 499–530 (1966).
- Thom M, Mathern GW, Cross JH, Bertram EH. Mesial temporal lobe epilepsy: how do we improve surgical outcome? Ann. Neurol. 68(4), 424–434 (2010).
- Wieser HG; ILAE Commission on Neurosurgery of Epilepsy. ILAE Commission Report. Mesial temporal lobe epilepsy with hippocampal sclerosis. Epilepsia 45(6), 695–714 (2004).
- Lin JJ, Salamon N, Lee AD et al. Reduced neocortical thickness and complexity mapped in mesial temporal lobe epilepsy with hippocampal sclerosis. Cereb. Cortex 17(9), 2007–2018 (2007).
- Salanova V, Markand O, Worth R. Temporal lobe epilepsy: analysis of patients with dual pathology. Acta Neurol. Scand. 109(2), 126–131 (2004).
- Bartoli A, Vulliemoz S, Haller S, Schaller K, Seeck M. Imaging techniques for presurgical evaluation of temporal lobe epilepsy. Imaging Med. 4(4), 443–459 (2012). & Reviews current literature on imaging modalities used for presurgical assessment of focal temporal lobe epilepsy.
- Von Oertzen J, Urbach H, Jungbluth S et al. Standard magnetic resonance imaging is inadequate for patients with refractory focal epilepsy. J. Neurol. Neurosurg. Psychiatry 73(6), 643–647 (2002).
- Hauptman JS, Salamon N, Mathern GW. Neuroimaging in the definition and organization of the epilepsies: we’re not there yet. Epilepsia 53(Suppl. 2), S22–S27 (2012).
- Recommendations for neuroimaging of patients with epilepsy. Commission on neuroimaging of the international league against epilepsy. Epilepsia 38(11), 1255–1256 (1997).
- Diehl B, Najm I, Mohamed A et al. Fluidattenuated inversion recovery: correlations of hippocampal cell densities with signal abnormalities. Neurology 57(6), 1029–1032 (2001).
- Rugg-Gunn FJ, Boulby PA, Symms MR, Barker GJ, Duncan JS. Whole-brain T2 mapping demonstrates occult abnormalities in focal epilepsy. Neurology 64(2), 318–325 (2005).
- Van Paesschen W, Revesz T, Duncan JS, King MD, Connelly A. Quantitative neuropathology and quantitative magnetic resonance imaging of the hippocampus in temporal lobe epilepsy. Ann. Neurol. 42(5), 756–766 (1997).
- Briellmann RS, Kalnins RM, Berkovic SF, Jackson GD. Hippocampal pathology in refractory temporal lobe epilepsy: T2- weighted signal change reflects dentate gliosis. Neurology 58(2), 265–271 (2002).
- Eriksson SH, Free SL, Thom M et al. Correlation of quantitative MRI and neuropathology in epilepsy surgical resection specimens – T2 correlates with neuronal tissue in gray matter. Neuroimage 37(1), 48–55 (2007).
- Lockwood-Estrin G, Thom M, Focke NK et al. Correlating 3T MRI and histopathology in patients undergoing epilepsy surgery. J. Neurosci. Methods 205(1), 182–189 (2012).
- Ashburner J, Friston KJ. Voxel-based morphometry – the methods. Neuroimage 11(6 Pt 1), 805–821 (2000).
- Mechelli A, Price CJ, Friston KJ, Ashburner J. Voxel-based morphometry of the human brain: methods and applications. Curr. Med. Imaging Rev. 1(2), 105–113 (2005).
- Whitwell JL. Voxel-based morphometry: an automated technique for assessing structural changes in the brain. J. Neurosci. 29(31), 9661–9664 (2009).
- Good CD, Johnsrude IS, Ashburner J, Henson RN, Friston KJ, Frackowiak RS. A voxel-based morphometric study of ageing in 465 normal adult human brains. Neuroimage 14(1 Pt 1), 21–36 (2001).
- Clarkson MJ, Cardoso MJ, Ridgway GR et al. A comparison of voxel and surface based cortical thickness estimation methods. Neuroimage 57(3), 856–865 (2011).
- Greve DN. An absolute beginner’s guide to surface- and voxel-based morphometric analysis. Presented at: ISMRM 19th Annual Meeting & Exhibition. Montréal, Québec, Canada, 7–13 May 2011.
- Keller SS, Roberts N. Voxel-based morphometry of temporal lobe epilepsy: an introduction and review of the literature. Epilepsia 49(5), 741–757 (2008). & Explains the voxel-based morphometry technique and reviews its application in temporal lobe epilepsy.
- Karas GB, Burton EJ, Rombouts SA et al. A comprehensive study of gray matter loss in patients with Alzheimer’s disease using optimized voxel-based morphometry. Neuroimage 18(4), 895–907 (2003).
- Price S, Paviour D, Scahill R et al. Voxelbased morphometry detects patterns of atrophy that help differentiate progressive supranuclear palsy and Parkinson’s disease. Neuroimage 23(2), 663–669 (2004).
- Voets NL, Hough MG, Douaud G et al. Evidence for abnormalities of cortical development in adolescent-onset schizophrenia. Neuroimage 43(4), 665–675 (2008).
- Keller SS, Mackay CE, Barrick TR, Wieshmann UC, Howard MA, Roberts N. Voxel-based morphometric comparison of hippocampal and extrahippocampal abnormalities in patients with left and right hippocampal atrophy. Neuroimage 16(1), 23–31 (2002).
- Bernasconi N, Duchesne S, Janke A, Lerch J, Collins DL, Bernasconi A. Whole-brain voxel-based statistical analysis of gray matter and white matter in temporal lobe epilepsy. Neuroimage 23(2), 717–723 (2004).
- Bonilha L, Rorden C, Castellano G et al. Voxel-based morphometry reveals gray matter network atrophy in refractory medial temporal lobe epilepsy. Arch. Neurol. 61(9), 1379–1384 (2004).
- McMillan AB, Hermann BP, Johnson SC, Hansen RR, Seidenberg M, Meyerand ME. Voxel-based morphometry of unilateral temporal lobe epilepsy reveals abnormalities in cerebral white matter. Neuroimage 23(1), 167–174 (2004).
- Keller SS, Wilke M, Wieshmann UC, Sluming VA, Roberts N. Comparison of standard and optimized voxel-based morphometry for analysis of brain changes associated with temporal lobe epilepsy. Neuroimage 23(3), 860–868 (2004).
- Pell GS, Briellmann RS, Chan CH, Pardoe H, Abbott DF, Jackson GD. Selection of the control group for VBM analysis: influence of covariates, matching and sample size. Neuroimage 41(4), 1324–1335 (2008).
- Mueller SG, Laxer KD, Cashdollar N, Buckley S, Paul C, Weiner MW. Voxelbased optimized morphometry (VBM) of gray and white matter in temporal lobe epilepsy (TLE) with and without mesial temporal sclerosis. Epilepsia 47(5), 900–907 (2006).
- Labate A, Cerasa A, Aguglia U, Mumoli L, Quattrone A, Gambardella A. Neocortical thinning in “benign” mesial temporal lobe epilepsy. Epilepsia 52(4), 712–717 (2011).
- Pail M, Brazdil M, Marecek R, Mikl M. An optimized voxel-based morphometric study of gray matter changes in patients with left-sided and right-sided mesial temporal lobe epilepsy and hippocampal sclerosis (MTLE/HS). Epilepsia 51(4), 511–518 (2010).
- Riederer F, Lanzenberger R, Kaya M, Prayer D, Serles W, Baumgartner C. Network atrophy in temporal lobe epilepsy: a voxel-based morphometry study. Neurology 71(6), 419–425 (2008).
- Santana MT, Jackowski AP, da Silva HH et al. Auras and clinical features in temporal lobe epilepsy: a new approach on the basis of voxel-based morphometry. Epilepsy Res. 89(2–3), 327–338 (2010).
- Lu J, Li W, He H, Feng F, Jin Z, Wu L. Altered hemispheric symmetry found in leftsided mesial temporal lobe epilepsy with hippocampal sclerosis (MTLE/HS) but not found in right-sided MTLE/HS. Magn. Reson. Imaging 31(1), 53–59 (2013).
- Pell GS, Briellmann RS, Pardoe H, Abbott DF, Jackson GD. Composite voxel-based analysis of volume and T2 relaxometry in temporal lobe epilepsy. Neuroimage 39(3), 1151–1161 (2008).
- Yu A, Li K, Li L, Shan B, Wang Y, Xue S. Whole-brain voxel-based morphometry of white matter in medial temporal lobe epilepsy. Eur. J. Radiol. 65(1), 86–90 (2008).
- Braga B, Yasuda CL, Cendes F. White matter atrophy in patients with mesial temporal lobe epilepsy: voxel-based morphometry analysis of T1- and T2-weighted MR images. Radiol. Res. Pract. 2012, 481378 (2012).
- Keller SS, Cresswell P, Denby C et al. Persistent seizures following left temporal lobe surgery are associated with posterior and bilateral structural and functional brain abnormalities. Epilepsy Res. 74(2–3), 131–139 (2007).
- Fischl B, Dale AM. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc. Natl Acad. Sci. USA 97(20), 11050–11055 (2000). & Presents an automated surface-based method for measuring the thickness of the cerebral cortex across the entire brain (i.e., FreeSurfer software, Harvard Martinos Center for Biomedical Imaging, MA, USA).
- Fischl B, Salat DH, Busa E et al. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron 33(3), 341–355 (2002).
- Fischl B, Salat DH, van der Kouwe AJ et al. Sequence-independent segmentation of magnetic resonance images. Neuroimage 23(Suppl. 1), S69–S84 (2004).
- Tosun D, Rettmann ME, Han X et al. Cortical surface segmentation and mapping. Neuroimage 23(Suppl. 1), S108–S118 (2004).
- Hurdal MK, Stephenson K. Cortical cartography using the discrete conformal approach of circle packings. Neuroimage 23(Suppl. 1), S119–S128 (2004).
- Akhondi-Asl A, Jafari-Khouzani K, Elisevich K, Soltanian-Zadeh H. Hippocampal volumetry for lateralization of temporal lobe epilepsy: automated versus manual methods. Neuroimage 54(Suppl. 1), S218–S226 (2011).
- Kim H, Mansi T, Bernasconi N, Bernasconi A. Surface-based multi-template automated hippocampal segmentation: application to temporal lobe epilepsy. Med. Image Anal. 16(7), 1445–1455 (2012).
- McDonald CR, Hagler DJ Jr, Ahmadi ME et al. Subcortical and cerebellar atrophy in mesial temporal lobe epilepsy revealed by automatic segmentation. Epilepsy Res. 79(2–3), 130–138 (2008).
- Alhusaini S, Doherty CP, Scanlon C et al. A cross-sectional MRI study of brain regional atrophy and clinical characteristics of temporal lobe epilepsy with hippocampal sclerosis. Epilepsy Res. 99(1–2), 156–166 (2012).
- Pardoe HR, Pell GS, Abbott DF, Jackson GD. Hippocampal volume assessment in temporal lobe epilepsy: how good is automated segmentation? Epilepsia 50(12), 2586–2592 (2009).
- Kim H, Chupin M, Colliot O, Bernhardt BC, Bernasconi N, Bernasconi A. Automatic hippocampal segmentation in temporal lobe epilepsy: impact of developmental abnormalities. Neuroimage 59(4), 3178–3186 (2012).
- Mueller SG, Laxer KD, Barakos J, Cheong I, Garcia P, Weiner MW. Widespread neocortical abnormalities in temporal lobe epilepsy with and without mesial sclerosis. Neuroimage 46(2), 353–359 (2009).
- McDonald CR, Hagler DJ, Ahmadi ME et al. Regional neocortical thinning in mesial temporal lobe epilepsy. Epilepsia 49(5), 794–803 (2008).
- McDonald CR, Taylor J, Hamberger M, Helmstaedter C, Hermann BP, Schefft B. Future directions in the neuropsychology of epilepsy. Epilepsy Behav. 22(1), 69–76 (2011). & Multimodal imaging for measuring subcortical volumes and cortical surface features in temporal lobe epilepsy.
- Mueller SG, Laxer KD, Barakos J et al. Involvement of the thalamocortical network in TLE with and without mesiotemporal sclerosis. Epilepsia 51(8), 1436–1445 (2010).
- Bernhardt BC, Bernasconi N, Kim H, Bernasconi A. Mapping thalamocortical network pathology in temporal lobe epilepsy. Neurology 78(2), 129–136 (2012).
- Bernhardt BC, Bernasconi N, Concha L, Bernasconi A. Cortical thickness analysis in temporal lobe epilepsy: reproducibility and relation to outcome. Neurology 74(22), 1776–1784 (2010).
- Thompson PM, Hayashi KM, Sowell ER et al. Mapping cortical change in Alzheimer’s disease, brain development, and schizophrenia. Neuroimage 23(Suppl. 1), S2–S18 (2004). & Presents a cortical pattern-matching brain mapping algorithm and its application in Alzheimer’s disease, schizophrenia, normal aging and abnormal brain development.
- Grenander U, Miller MI. Computational anatomy: an emerging discipline. Quart. App. Math. 56(4), 617–694 (1998).
- Thompson PM, Hayashi KM, De Zubicaray GI et al. Mapping hippocampal and ventricular change in Alzheimer disease. Neuroimage 22(4), 1754–1766 (2004).
- Hogan RE, Carne RP, Kilpatrick CJ et al. Hippocampal deformation mapping in MRI negative PET positive temporal lobe epilepsy. J. Neurol. Neurosurg. Psychiatry 79(6), 636–640 (2008).
- Ogren JA, Bragin A, Wilson CL et al. Three-dimensional hippocampal atrophy maps distinguish two common temporal lobe seizure-onset patterns. Epilepsia 50(6), 1361–1370 (2009). & Shows 3D hippocampal atrophy maps, produced by a cortical pattern-matching technique, distinguishing between two common temporal lobe seizure-onset patterns.
- Lin JJ, Salamon N, Dutton RA et al. Three-dimensional preoperative maps of hippocampal atrophy predict surgical outcomes in temporal lobe epilepsy. Neurology 65(7), 1094–1097 (2005).
- Zeineh MM, Engel SA, Bookheimer SY. Application of cortical unfolding techniques to functional MRI of the human hippocampal region. Neuroimage 11(6 Pt 1), 668–683 (2000).
- Ekstrom AD, Bazih AJ, Suthana NA et al. Advances in high-resolution imaging and computational unfolding of the human hippocampus. Neuroimage 47(1), 42–49 (2009).
- Ekstrom A, Suthana N, Millett D, Fried I, Bookheimer S. Correlation between BOLD fMRI and t-band local field potentials in the human hippocampal area. J. Neurophysiol. 101(5), 2668–2678 (2009).
- Staba RJ, Ekstrom AD, Suthana NA et al. Gray matter loss correlates with mesial temporal lobe neuronal hyperexcitability inside the human seizure-onset zone. Epilepsia 53(1), 25–34 (2012).
- Gaonkar B, Pohl K, Davatzikos C. Pattern based morphometry. Med. Image Comput. Comput. Assist. Interv. 14(Pt 2), 459–466 (2011). & Details of a pattern-based morphometry algorithm, designed to address the shortcomings of voxel-based morphometry techniques.
- Toews M, Wells W 3rd, Collins DL, Arbel T. Feature-based morphometry: discovering group-related anatomical patterns. Neuroimage 49(3), 2318–2327 (2010).
- Mangin JF, Riviere D, Cachia A et al. Object-based morphometry of the cerebral cortex. IEEE Trans. Med. Imaging 23(8), 968–982 (2004).
- Mangin JF, Rivière D, Cachia A et al. Objectbased strategy for morphometry of the cerebral cortex. Inf. Process. Med. Imaging 18, 160–171 (2003).
- Wiebe S, Blume WT, Girvin JP, Eliasziw M; Effectiveness, Efficiency of Surgery for Temporal Lobe Epilepsy Study G. A randomized, controlled trial of surgery for temporal-lobe epilepsy. N. Engl. J. Med. 345(5), 311–318 (2001).
- Rugg-Gunn FJ, Eriksson SH, Boulby PA, Symms MR, Barker GJ, Duncan JS. Magnetization transfer imaging in focal epilepsy. Neurology 60(10), 1638–1645 (2003).
- Morimoto E, Kanagaki M, Okada T et al. Anterior temporal lobe white matter abnormal signal (ATLAS) as an indicator of seizure focus laterality in temporal lobe epilepsy: comparison of double inversion recovery, FLAIR and T2W MR imaging. Eur. Radiol. 23(1), 3–11 (2013).
- Koepp MJ, Woermann FG. Imaging structure and function in refractory focal epilepsy. Lancet Neurol. 4(1), 42–53 (2005). & Reviews structual and functional imaging modalities used in studies of focal refractory epilepsy.
- Yoo SY, Chang KH, Song IC et al. Apparent diffusion coefficient value of the hippocampus in patients with hippocampal sclerosis and in healthy volunteers. AJNR Am. J. Neuroradiol. 23(5), 809–812 (2002).
- Kantarci K, Shin C, Britton JW, So EL, Cascino GD, Jack CR Jr. Comparative diagnostic utility of 1H MRS and DWI in evaluation of temporal lobe epilepsy. Neurology 58(12), 1745–1753 (2002).
- Lee JH, Chung CK, Song IC, Chang KH, Kim HJ. Limited utility of interictal apparent diffusion coefficient in the evaluation of hippocampal sclerosis. Acta Neurol. Scand. 110(1), 53–58 (2004).
- O’Brien TJ, David EP, Kilpatrick CJ, Desmond P, Tress B. Contrast-enhanced perfusion and diffusion MRI accurately lateralize temporal lobe epilepsy: a pilot study. J. Clin. Neurosci. 14(9), 841–849 (2007).
- O’Dwyer R, Wehner T, Lapresto E et al. Differences in corpus callosum volume and diffusivity between temporal and frontal lobe epilepsy. Epilepsy Behav. 19(3), 376–382 (2010).
- Londono A, Castillo M, Lee YZ, Smith JK. Apparent diffusion coefficient measurements in the hippocampi in patients with temporal lobe seizures. AJNR Am. J. Neuroradiol. 24(8), 1582–1586 (2003).
- Salmenpera TM, Symms MR, Rugg-Gunn FJ et al. Evaluation of quantitative magnetic resonance imaging contrasts in MRI-negative refractory focal epilepsy. Epilepsia 48(2), 229–237 (2007).
- Wehner T, Lapresto E, Tkach J et al. The value of interictal diffusion-weighted imaging in lateralizing temporal lobe epilepsy. Neurology 68(2), 122–127 (2007).
- Powell HW, Guye M, Parker GJ et al. Noninvasive in vivo demonstration of the connections of the human parahippocampal gyrus. Neuroimage 22(2), 740–747 (2004).
- Assaf BA, Mohamed FB, Abou-Khaled KJ et al. Diffusion tensor imaging of the hippocampal formation in temporal lobeepilepsy. AJNR Am. J. Neuroradiol. 24(9), 1857–1862 (2003).
- Shon YM, Kim YI, Koo BB et al. Groupspecific regional white matter abnormality revealed in diffusion tensor imaging of medial temporal lobe epilepsy without hippocampal sclerosis. Epilepsia 51(4), 529–535 (2010).
- Keller SS, Ahrens T, Mohammadi S et al. Voxel-based statistical analysis of fractional anisotropy and mean diffusivity in patients with unilateral temporal lobe epilepsy of unknown cause. J. Neuroimaging doi:10.1111/j.1552-6569.2011.00673.x (2011) (Epub ahead of print).
- Focke NK, Yogarajah M, Bonelli SB, Bartlett PA, Symms MR, Duncan JS. Voxel-based diffusion tensor imaging in patients with mesial temporal lobe epilepsy and hippocampal sclerosis. Neuroimage 40(2), 728–737 (2008).
- Knake S, Salat DH, Halgren E, Halko MA, Greve DN, Grant PE. Changes in white matter microstructure in patients with TLE and hippocampal sclerosis. Epileptic Disord. 11(3), 244–250 (2009).
- Afzali M, Soltanian-Zadeh H, Elisevich KV. Tract based spatial statistical analysis and voxel based morphometry of diffusion indices in temporal lobe epilepsy. Comput. Biol. Med. 41(12), 1082–1091 (2011).
- Hagler DJ Jr, Ahmadi ME, Kuperman J et al. Automated white-matter tractography using a probabilistic diffusion tensor atlas: application to temporal lobe epilepsy. Hum. Brain Mapp. 30(5), 1535–1547 (2009).
- Keller SS, Schoene-Bake JC, Gerdes JS, Weber B, Deppe M. Concomitant fractional anisotropy and volumetric abnormalities in temporal lobe epilepsy: cross-sectional evidence for progressive neurologic injury. PLoS ONE 7(10), e46791 (2012).
- Duning T, Kellinghaus C, Mohammadi S et al. Individual white matter fractional anisotropy analysis on patients with MRI negative partial epilepsy. J. Neurol. Neurosurg. Psychiatry 81(2), 136–139 (2010).
- Concha L, Kim H, Bernasconi A, Bernhardt BC, Bernasconi N. Spatial patterns of water diffusion along white matter tracts in temporal lobe epilepsy. Neurology 79(5), 455–462 (2012).
- Liacu D, Idy-Peretti I, Ducreux D, Bouilleret V, de Marco G. Diffusion tensor imaging tractography parameters of limbic system bundles in temporal lobe epilepsy patients. J. Magn. Reson. Imaging 36(3), 561–568 (2012).
- Yogarajah M, Focke NK, Bonelli SB et al. The structural plasticity of white matter networks following anterior temporal lobe resection. Brain 133(Pt 8), 2348–2364 (2010).
- Nguyen D, Vargas MI, Khaw N et al. Diffusion tensor imaging analysis with tract-based spatial statistics of the white matter abnormalities after epilepsy surgery. Epilepsy Res. doi:10.1016/j.eplepsyres. 2011.02.001 (2011) (Epub ahead of print).
- Focke NK, Yogarajah M, Symms MR, Gruber O, Paulus W, Duncan JS. Automated MR image classification in temporal lobe epilepsy. Neuroimage 59(1), 356–362 (2012).
- Duchesne S, Bernasconi N, Bernasconi A, Collins DL. MR-based neurological disease classification methodology: application to lateralization of seizure focus in temporal lobe epilepsy. Neuroimage 29(2), 557–566 (2006).
- Urbach H, Siebenhaar G, Koenig R et al. Limbic system abnormalities associated with Ammon’s horn sclerosis do not alter seizure outcome after amygdalohippocampectomy. Epilepsia 46(4), 549–555 (2005).
- Bonilha L, Elm JJ, Edwards JC et al. How common is brain atrophy in patients with medial temporal lobe epilepsy? Epilepsia 51(9), 1774–1779 (2010).
- Cormack F, Gadian DG, Vargha-Khadem F, Cross JH, Connelly A, Baldeweg T. Extrahippocampal grey matter density abnormalities in paediatric mesial temporal sclerosis. Neuroimage 27(3), 635–643 (2005).
- Davatzikos C. Why voxel-based morphometric analysis should be used with great caution when characterizing group differences. Neuroimage 23(1), 17–20 (2004).
- Scanlon C, Mueller SG, Tosun D et al. Impact of methodologic choice for automatic detection of different aspects of brain atrophy by using temporal lobe epilepsy as a model. AJNR Am. J. Neuroradiol. 32(9), 1669–1676 (2011).
- Mehta S, Grabowski TJ, Trivedi Y, Damasio H. Evaluation of voxel-based morphometry for focal lesion detection in individuals. Neuroimage 20(3), 1438–1454 (2003).
- Scarpazza C, Sartori G, De Simone MS, Mechelli A. When the single matters more than the group: very high false positive rates in single case Voxel Based Morphometry. Neuroimage 70, 175–188 (2013).
- Woermann FG, Free SL, Koepp MJ, Ashburner J, Duncan JS. Voxel-by-voxel comparison of automatically segmented cerebral gray matter – a rater-independent comparison of structural MRI in patients with epilepsy. Neuroimage 10(4), 373–384 (1999).
- Rezaie MG, Soltanian-Zadeh H, Siadat MR, Elisevich KV. Soft computing approaches to computer aided decision making for temporal lobe epilepsy. Presented at: Annual Meeting of the North American Fuzzy Information Processing Society (NAFIPS). Ann Arbor, MI, USA, 26–28 June 2005.
- Fakhraei S, Soltanianzadeh H, Jafari- Khouzani K, Elisevich K, Fotouhi F. Confident surgical decision making in temporal lobe epilepsy by heterogeneous classifier ensembles. Proceedings of the IEEE 11th International Conference Data Mining Workshops (ICDMW). Vancouver, BC, Canada, 11 December 2011.
- Bonilha L, Nesland T, Martz GU et al. Medial temporal lobe epilepsy is associated with neuronal fibre loss and paradoxical increase in structural connectivity of limbic structures. J. Neurol. Neurosurg. Psychiatry 83(9), 903–909 (2012).
- Bernhardt BC, Chen Z, He Y, Evans AC, Bernasconi N. Graph-theoretical analysis reveals disrupted small-world organization of cortical thickness correlation networks in temporal lobe epilepsy. Cereb. Cortex 21(9), 2147–2157 (2011).
- Mueller SG, Laxer KD, Schuff N, Weiner MW. Voxel-based T2 relaxation rate measurements in temporal lobe epilepsy (TLE) with and without mesial temporal sclerosis. Epilepsia 48(2), 220–228 (2007).
- Ogren JA, Wilson CL, Bragin A et al. Threedimensional surface maps link local atrophy and fast ripples in human epileptic hippocampus. Ann. Neurol. 66(6), 783–791 (2009).
- Spencer S, Huh L. Outcomes of epilepsy surgery in adults and children. Lancet Neurol. 7(6), 525–537 (2008).
- Blumcke I, Pauli E, Clusmann H et al. A new clinico-pathological classification system for mesial temporal sclerosis. Acta Neuropathol. 113(3), 235–244 (2007).
- Engel J Jr, McDermott MP, Wiebe S et al. Early surgical therapy for drug-resistant temporal lobe epilepsy: a randomized trial. JAMA 307(9), 922–930 (2012).
- Salamon N, Kung J, Shaw SJ et al. FDGPET/ MRI coregistration improves detection of cortical dysplasia in patients with epilepsy. Neurology 71(20), 1594–1601 (2008).
- Willmann O, Wennberg R, May T, Woermann FG, Pohlmann-Eden B. The contribution of 18F-FDG PET in preoperative epilepsy surgery evaluation for patients with temporal lobe epilepsy. A meta-analysis. Seizure 16(6), 509–520 (2007).
- Knowlton RC. The role of FDG-PET, ictal SPECT, and MEG in the epilepsy surgery evaluation. Epilepsy Behav. 8(1), 91–101 (2006).
- Worrell GA. Imaging focal cortical dysplasia in refractory epilepsy. Epilepsy Curr. 12(1), 35–36 (2012). & Discusses the sensitivity of several neuroimaging tools in identifying focal cortical dysplasia (a common cause of drug-resistant epilepsy).
- Rathore C, Kesavadas C, Ajith J, Sasikala A, Sarma SP, Radhakrishnan K. Cost-effective utilization of single photon emission computed tomography (SPECT) in decision making for epilepsy surgery. Seizure 20(2), 107–114 (2011).
- Haneef Z, Lenartowicz A, Yeh HJ, Engel J Jr, Stern JM. Effect of lateralized temporal lobe epilepsy on the default mode network. Epilepsy Behav. 25(3), 350–357 (2012).
- Vulliemoz S, Rodionov R, Carmichael DW et al. Continuous EEG source imaging enhances analysis of EEG-fMRI in focal epilepsy. Neuroimage 49(4), 3219–3229 (2010).
- De Tiège X, Connelly A, Liégeois F et al. Influence of motor functional magnetic resonance imaging on the surgical management of children and adolescents with symptomatic focal epilepsy. Neurosurgery 64(5), 856–864; discussion 864 (2009).
- Zhang J, Mei S, Liu Q et al. fMRI and DTI assessment of patients undergoing radical epilepsy surgery. Epilepsy Res. 104(3), 253–263 (2013).
- De Tiege X, Carrette E, Legros B et al. Clinical added value of magnetic source imaging in the presurgical evaluation of refractory focal epilepsy. J. Neurol. Neurosurg. Psychiatry 83(4), 417–423 (2012).
- Leijten FS, Huiskamp G. Interictal electromagnetic source imaging in focal epilepsy. Practices, results and recommendations. Curr. Opin. Neurol. 21(4), 437–445 (2008).
- Richardson MP. Large scale brain models of epilepsy. dynamics meets connectomics. J. Neurol. Neurosurg. Psychiatry 83(12), 1238–1248 (2012).
- Engel J Jr, Thompson PM, Stern JM et al. Connectomics and epilepsy. Curr. Opin. Neurol. 26(2), 186-194 (2013).
- Turken AU, Herron TJ, Kang X et al. Multimodal surface-based morphometry reveals diffuse cortical atrophy in traumatic brain injury. BMC Med. Imaging 9, 20 (2009).