Research Article - Clinical Practice (2021) Volume 18, Issue 3
Resistant Bacteria in Children with Community-Acquired Febrile Illness in a Tertiary Hospital in Nigeria
- Corresponding Author:
- Shittu Abdulwahab Adewale
Al-Noor Specialist Hospital
Makkah Al-Mukarramah, Makkah
Kingdom of Saudi Arabia
E-mail: abumaryamshittu@gmail.com
Abstract
Background: Blood bacterial infection is a cause of serious illness in children, especially the antibiotic resistant organisms. Since there is variation in the causative organisms with different location, there is need to determine the burden in our location aside the low data recorded. This study determined prevalence of bacteraemia, resistance pattern of implicated organisms and the role of Procalcitonin (PCT) in children with Community Acquired Bacteraemia (CAB) at a tertiary hospital in Nigeria. It is to enhance focus on antibiotic stewardship in clinical practice. Methods: Children clinically suspected to have bacteraemia at presentation during 13 months of the period of study were recruited. Their blood samples were cultured and assayed for serum procalcitonin. Antibiotic resistance was determined on isolated bacteria, and polymerase chain reaction was used to confirm implicated genes. The data generated were analyzed using appropriate descriptive and inferential statistics. Results: A total of 343 children ≤ 14-years were evaluated, and 94 (27.4%) had bacteraemia. The most common organisms were Staphylococcus aureus (n=66; 70.2%), Stenotrophomonas maltophilia (n=7; 7.4%), and coagulase negative staphylococci (n=6; 6.4%). More than fifty percent of all the isolates were multidrug-resistant. Twenty-one of 21 Staphylococcus aureus had mecA gene and three Gram-negative isolates had at least one of blaCTX-M/blaSHV/blaTEM genes. Elevated serum procalcitonin level was significantly associated with bacteraemia (χ²=21.652, ρ =<0.001), sensitivity of 83.0% and negative predictive value of 87.3% (80.4-92.0). Congenital malformation was most associated with community acquired bacteraemia. Conclusion: Nearly 30% of children suspected with bacteraemia in the studied population had positive blood-culture. The most isolated pathogen was Staphylococcus aureus; and a third of the pathogens were multi-drug resistant. Procalcitonin assay is useful in excluding bacteraemia in febrile children. Resistant bacteria pathogens are not uncommon in the community.
Keywords
multi-drug resistance, extended spectrum beta-lactamase, meca genet
Introduction
Bacterial infections are common in children and are a serious cause of childhood mortality in sub-Saharan Africa [1-3]. Community Acquired Bacteremia (CAB) is due to dissemination of viable bacterial in the bloodstream from a primarily infected site, of an individual outside the period of hospital admission [1]. The diagnosis is sometimes obscured, in this environment, due to other endemic illnesses like malaria [2,4]. The gold standard for diagnosis of bacteremia is by blood culture [5]. The diagnosis and care of patients with bacteremia can be enhanced by biomarkers like Procalcitonin (PCT) [6]. Information about CAB in southwestern region of Nigeria is sparse [1-3,7- 9]. Immediate treatment of CAB may increase patients’ chance of survival, whereas delay may increase the morbidity and mortality [10]. In most cases, bacteremia is due to infections of the respiratory, gastrointestinal tracts, central nervous system and soft-tissues. The initial site of infection may be obscured (primary bacteremia), despite testing with the best tests available [11]. It is especially so in malnourished or HIV-infected children [1-3,8,9]. Other risk factors for CAB are age, malaria, low birth weight, overcrowding, deficiencies of minerals and vitamins, inadequate immunization, breast feeding, adverse obstetrics history, congenital anomaly, poor socio-economic status, and care givers’ inadequate health education [1,3,8,12- 17]. Commonly implicated organisms are Staphylococcus aureus, Streptococcus pneumonia, Haemophilus influenzae, Klebsiella pneumoniae and Salmonella typhi [3,8,14,18,19]. Asides low data on epidemiologic patterns of CAB in our location, the emergence and dissemination of multidrug-resistant bacteria causing difficult-totreat infections, requires attention [1,3,8,9].
The earlier studies done in our location of study showed that the leading cause of bacteremia in young children is Gram negative bacteria in Ilesa [12], while Ile-Ife documented Gram positive bacteria [13,20,21]. These isolated organisms were sensitive to the commonly used antibiotics then, this included quinolones and third-generation cephalosporin [20,21] although one of the studies from Ile-Ife detected in vitro resistance some of the isolates [13]. With the increasing incidence of community acquired resistant bacteria pathogens, there could have been alterations in what was documented. Besides, a regular surveillance is important in determining the epidemiologic pattern of bacteria pathogens to maintain a rational antibiotic use. This will also assist in reducing morbidity and mortality, associated with community acquired bacteremia. As at the time of this study initial antibiotics chosen for the treatment of bacteremia-related cases in this locality is mainly the combination of gentamicin with ceftriaxone or ampicillin for new born and infants; while in older children, it is cefuroxime with or without gentamicin.
The aim of this study was to determine the prevalence of bacteremia, resistance pattern of the causative organisms and role of procalcitonin in children with CAB seen at a university teaching hospital in southwestern Nigeria.
Methodology
■ Study Site
This study was conducted at Obafemi Awolowo University Teaching Hospitals Complex (OAUTHC), a tertiary hospital in Ile-Ife, Osun State, Nigeria. Osun State is situated in the tropical rain forest zone, covering an area of approximately 14,875 sq km and lies between latitude 7˚30′0″ N and longitude 4˚30′0″ E. It provides services through six health care units, two of which have inpatient services-Ile-Ife Hospital Unit and the Wesley Guild Hospital with 535 and 212 bed complements respectively. Approval for the study (Approval number- IRB/IEC/0004553 and Protocol number- ERC/2015/04/06) was obtained from the Ethical Review Committee of OAUTHC. The patients were recruited between August 2015 and August 2016 at the children emergency and neonatal wards.
■ Sampling
All patients with febrile illness suspected of having a community acquired bacteremia, based on pediatrician assessment, were recruited. Children with diagnosis not related to sepsis, those admitted for more than 48 hours before developing fever, children who were discharged from hospital and re-presented with fever within 48 hours and children whose parent (s) or guardian (s) did not give consent were all excluded from the study. Demographic information and clinical details of each patient was recorded on forms specific for this study. The venipuncture site was cleaned with 70% alcohol and povidone-Iodine, then one to three milliliters of venous blood were drawn and introduced aseptically into two blood culture bottles (by BD PeadPlus bottle; Becton Dickinson). Also, three drops of plasma were added into the well of procalcitonin rapid diagnostic kit (StrongStep® PCT Rapid Test).
■ Processing
The PCT rapid kit was read after 15 minutes and interpreted based on the manufacturer recommendation. Readings >0.5 were taken as a case of sepsis. The blood culture samples were incubated using the BD Bactec™ 9050 Blood Culture System; Becton Dickinson. The Gram-negative organisms were identified with morphology, microscopy and Microbact GNB 24E, identification kit by Oxoid, while Gram-positive organisms were identified with morphology, microscopy, catalase, coagulase and optochin testing. Susceptibility testing was done using the following antibiotic discs: ampicillin (10 μg), ceftriaxone (30 μg), ceftazidime (30 μg), cefotaxime (30 μg), cefepime (30 μg), co-amoxiclav (20/10 μg), cefuroxime (30 μg), ciprofloxacin (5 μg), gentamicin (10 μg), imipenem (10 μg), levofloxacin (5 μg), cotrimoxazole (1.25/23.75 μg), piperacillin/ tazobactam (110 μg), ampicillin/sulbactam (30 μg), aztreonam (30 μg) and azithromycin (15 μg) by modified Kirby-Bauer disk diffusion technique in line with the Clinical Laboratory Science Institute (CLSI) guidelines [22].
Phenotypically positive isolates for Extended Spectrum Beta-Lactamase (ESBL) producing bacteria and Methicillin-Resistant Staphylococcus aureus (MRSA) were subjected to molecular confirmation. The DNA extraction was conducted by the boiling method and the resulting DNA suspension was used as a template DNA for Polymerase Chain Reaction (PCR) based detection [23]. The ESBL genes tested for were blaTEM, blaSHV and blaCTX-M, while the MRSA gene tested for was mecA. The primers used, as shown in TABLE 1, were commercially synthesized by Inqaba biotechnical Industries (Pty) Ltd. (South Africa) [24-26]. The procedure was followed according to the specification of the manufacturer.
TABLE 1. Sets of primers used to identify ESBL and mecA genes.
| Genes | Primers | ||
|---|---|---|---|
| Forward | Reverse | Amplification at | |
| blaSHV | CGCCTGTGTATTATCTCCCT | CGAGTAGTCCACCAGATCCT | 293 bp region |
| blaTEM | TTTCGTGTCGCCCTTATTCC | ATCGTTGTCAGAAGTAAGTTGG | 403 bp region |
| blaCTX-M | CGCTGTTGTTAGGAAGTGTG | GGCTGGGTGAAGTAAGTGAC | 569 bp region |
| mecA | AGTTCTGCAGTACCGGATTTGC | ATCGATGGTAAAGGTTGGC | 530 bp region |
The PCR products were electrophoresed, and the amplified bands were visualized under ultraviolet light with an UVitech transilluminator (Aveburg, Cambridge UK). The position of the amplified product was estimated by the position of 100 base pair molecular weight marker (Bio Lab Scientific Ltd., Toronto, Canada). All the data generated were entered into Microsoft Excel and processed using Statistical Package for the Social Sciences (SPSS) version 20.
Results
Three hundred forty-three infants and children aged 1 hour to 14 years [Median (IQR); 5 wks (2 days-34 months)] were evaluated, male: female ratio was found to be 1.1:1.0. The percentage of subjects recruited decreased progressively with age, from 49.3% in neonates to 7.6% in adolescents. The most common diagnosis was sepsis 142 (44.8%) and other illnesses include soft-tissue infection 26 (8.2%) and pneumonia 23 (7.3%). Ninety-four (27.4%) of the 343 patients had proven bacteremia. The number of cases seen during the harmattan season was 60 which yielded 13.8% of the positive cases. While in the rainy season 283 cases yielded 86.2% of the total positive cases. The frequency of patients is higher in the younger age group, especially the neonates, but the fraction of those with bacteremia within the different groups is higher in the older children. The case fatality rate among recruited patients was 12.8%. There is was no significant association of bacteremia with gender, age, season, socio-economic class or final outcome the illness.
Seventy-eight of the 94 bacteremia subjects had positive serum procalcitonin assay for bacterial infection. Procalcitonin assay was significantly associated with bacteraemia (χ²=21.652, ρ=<0.001). The sensitivity is equal to 83.0% while the specificity is equal to 44.2%; also the positive predictive value and negative predictive values were 35.9% (29.9-42.5) and 87.3% (80.4- 92.0) respectively. The organisms isolated from patients’ blood cultures are listed in TABLE 2. Only 98 of the 343 blood culture pairs yielded the same organisms/pair. Ninety-four (27.4%) of these pairs were considered clinically significant community-acquired organisms while 4 (1.2%) were considered contaminants. Of the clinically significant organisms isolated, 75 (79.8%) were Gram-positive, and 19 (20.2%) were Gram-negative. The most prevalent organism was Staphylococcus aureus (70.2%), followed by Stenotrophomonas maltophilia (7.4%) and coagulase-negative staphylococci (CoNS; 6.4%). The organism Staphylococcus aureus was seen to be predominant in all age groups.
TABLE 2. Organisms isolated from blood cultures.
| Isolate | Frequency (%) | ||
|---|---|---|---|
| Staphylococcus aureus | 66 (70.2) | Gram-positives | Clinically significant organisms |
| Coagulase negative staphylococci | 6 (6.4) | ||
| Steptococcus pnaeumoniae | 2 (2.1) | ||
| Viridans group of streptococci | 1 (1.1) | ||
| Stenotrophomonas maltophilia | 7 (7.4) | Gram-negatives | |
| Acinetobacter baumanii | 2 (2.1) | ||
| Acinetobacter haemolyticus | 1 (1.1) | ||
| Proteus mirabilis | 1 (1.1) | ||
| Escherichia coli | 2 (2.1) | ||
| Citrobacter sedlaki | 1 (1.1) | ||
| Hafnia alvei | 2 (2.1) | ||
| Salmonella Arizonae | 1 (1.1) | ||
| Salmonella Typhi | 2 (2.1) | ||
| Total | 94 (100) | ||
| Gram-positive bacilli | 3 (75.0) | Contaminants | |
| Fungi | 1 (25.0) | ||
| Total | 4 (100) | ||
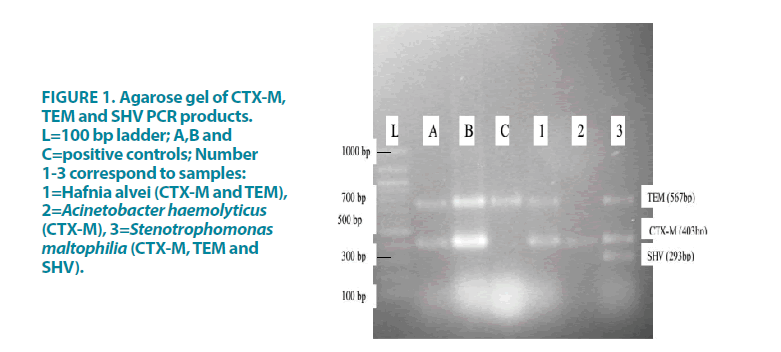
More than 50% of the isolates were resistant to at least three antibiotics from the different classes of antibiotics (multidrug-resistant), including the commonly used antibiotics [27]. TABLE 3 shows the antibiotic resistance patterns of the isolated Gram-negative bacteria. High rates of resistance were present in Stenotrophomonas maltophilia, Acinetobacter baumanii, Acinetobacter haemolyticus and Proteus mirabilis, whereas Salmonella Typhi, Salmonella Arizonae and Citrobacter sedlaki had the lowest rates of resistance. Three Gram-negative bacteria were phenotypically positive for ESBL production. Thirty-three percent of the Gramnegative isolates had ESBL genes as shown in FIGURE 1. The genes identified with PCR were blaSHV/TEM/CTX-M (in S. maltophilia), blaCTX-M/TEM (in H. alvei) and blaCTX-M (in A. haemolyticus).
TABLE 3. Resistance pattern of the isolated Gram-negative bacteria.
| Antibiotic | Isolates, n (%) resistant | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| S. maltophilia | A. baumannii. | A. haemolyticus | E. coli | P. mirabilis | C. sedlaki | H.alvei | S.enterica Arezonae | S.enterica Typhi | |
| (n=7) | (n=2) | (n=1) | (n=2) | (n=1) | (n=1) | (n=2) | (n=1) | (n=2) | |
| Cefoxitin | 2 (28.6) | 0 (0.0) | 1 (100.0) | 0 (0.0) | 1 (100.0) | 0 (0.0) | 1 (50.0) | 0 (0.0) | 0 (0.0) |
| Ceftriaxone | 3 (42.9) | 0 (0.0) | 1 (100.0) | 0 (0.0) | 1 (100.0) | 0 (0.0) | 1 (50.0) | 0 (0.0) | 0 (0.0) |
| Cefotaxime | 3 (42.9) | 0 (0.0) | 1 (100.0) | 1 (50.0) | 1 (100.0) | 0 (0.0) | 1 (50.0) | 0 (0.0) | 0 (0.0) |
| Ceftazidime | 2 (28.6) | 1 (50.0) | 1 (100.0) | 0 (0.0) | 1 (100.0) | 0 (0.0) | 1 (50.0) | 0 (0.0) | 0 (0.0) |
| Cefepime | 3 (42.9) | 1 (50.0) | 1 (100.0) | 0 (0.0) | 1 (100.0) | 0 (0.0) | 1 (50.0) | 0 (0.0) | 0 (0.0) |
| Aztreonam | 3 (42.9) | 0 (0.0) | 1 (100.0) | 0 (0.0) | 1 (100.0) | 0 (0.0) | 1 (50.0) | 0 (0.0) | 0 (0.0) |
| Gentamicin | 3 (42.9) | 1 (50.0) | 1 (100.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (50.0) | 0 (0.0) | 0 (0.0) |
| Amikacin | 1 (14.3) | 1 (50.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (50.0) | 0 (0.0) | 0 (0.0) |
| Ciprofloxacin | 3 (42.9) | 1 (50.0) | 1 (100.0) | 1 (50.0) | 1 (100.0) | 0 (0.0) | 1 (50.0) | 0 (0.0) | 0 (0.0) |
| Levofloxacin | 2 (28.6) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (100.0) | 0 (0.0) | 1 (50.0) | 0 (0.0) | 0 (0.0) |
| Tetracycline | 4 (57.1) | 2 (100.0) | 1 (100.0) | 2 (100.0) | 1 (100.0) | 0 (0.0) | 2 (100.0) | 0 (0.0) | 2 (100.0) |
| Imipenem | 2 (28.6) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (50.0) | 0 (0.0) | 0 (0.0) |
| Ampicilin | 6 (85.7) | 0 (0.0) | 1 (100.0) | 1 (50.0) | 1 (100.0) | 0 (0.0) | 2 (100.0) | 0 (0.0) | 1 (50.0) |
| Piperacillin/Tazobactam | 1 (14.3) | 0 (0.0) | 1 (100.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (50.0) | 0 (0.0) | 0 (0.0) |
| Co-Amoxiclav | 3 (42.9) | 0 (0.0) | 1 (100.0) | 1 (50.0) | 1 (100.0) | 0 (0.0) | 2 (100.0) | 0 (0.0) | 0 (0.0) |
| Ampicillin/Sulbactam | 3 (42.9) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (50.0) | 0 (0.0) | 0 (0.0) |
| Co-Trimoxazole | 5 (71.4) | 2 (100.0) | 0 (0.0) | 2 (100.0) | 1 (100.0) | 0 (0.0) | 2 (100.0) | 0 (0.0) | 2 (100.0) |
| Chloramphenicol | 5 (71.4) | 1 (50.0) | 1 (100.0) | 2 (100.0) | 1 (100.0) | 0 (0.0) | 2 (100.0) | 0 (0.0) | 2 (100.0) |
Figure 1: Agarose gel of CTX-M, TEM and SHV PCR products. L=100 bp ladder; A,B and C=positive controls; Number 1-3 correspond to samples: 1=Hafnia alvei (CTX-M and TEM), 2=Acinetobacter haemolyticus (CTX-M), 3=Stenotrophomonas maltophilia (CTX-M, TEM and SHV).
The antibiotic resistance pattern of Gram-positive isolates is shown in TABLE 4. Staphylococcus aureus had high rates of resistance to ampicillin, penicillin, cefuroxime, ceftriaxone, ceftazidime, cefepime, tetracycline, co-trimoxazole and azithromycin. Nineteen (28.8%) of the Gram positive bacteria identified as Staphylococcus aureus were Methicillin Resistant (MRSA). The six identified coagulase-negative staphylococci had a similar resistance pattern with 33.3% being methicillin-resistant coagulase-negative staphylococci (MR-CoNS).
Table 4. Resistance pattern of the isolated Gram-positive bacteria.
| Antibiotic | Isolates, n (%) resistant | |||
|---|---|---|---|---|
| S. aureus | CoNS | S. pneumonia | VGS | |
| (n=66) | (n=6) | (n=2) | (n=1) | |
| Cefuroxime | 38 (57.6) | 3 (50.0) | 0 (0.0) | 0 (0.0) |
| Cefoxitin | 19 (28.8) | 3 (50.0) | ||
| Ceftriaxone | 46 (69.7) | 5 (83.3) | 0 (0.0) | 0 (0.0) |
| Ceftazidime | 50 (75.8) | 6 (100.0) | 0 (0.0) | 0 (0.0) |
| Cefepime | 55 (83.3) | 4 (66.7) | 0 (0.0) | 0 (0.0) |
| Azithromycin | 38 (57.6) | 3 (50.0) | 0 (0.0) | 0 (0.0) |
| Erythromycin | 28 (42.4) | 3 (50.0) | 0 (0.0) | 0 (0.0) |
| Gentamicin | 24 (36.4) | 2 (33.3) | 0 (0.0) | 0 (0.0) |
| Amikacin | 7 (10.6) | 0 (0.0) | 0 (0.0) | 1 (100.0) |
| Ciprofloxacin | 18 (27.3) | 1 (16.7) | 0 (0.0) | 0 (0.0) |
| Levofloxacin | 15 (22.7) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Tetracycline | 44 (66.7) | 4 (66.7) | 1 (50.0) | 0 (0.0) |
| Ampicilin | 49 (74.2) | 4 (66.7) | 0 (0.0) | 0 (0.0) |
| Amoxicillin | 0 (0.0) | 0 (0.0) | ||
| Penicillin | 60 (90.9) | 5 (83.3) | 0 (0.0) | 0 (0.0) |
| Carbenicillin | 0 (0.0) | 0 (0.0) | ||
| Piperacillin/Tazobactam | 18 (27.3) | 2 (33.3) | 0 (0.0) | 0 (0.0) |
| Co-amoxiclav | 17 (25.8) | 1 (16.7) | 0 (0.0) | 0 (0.0) |
| Ampicillin/Sulbactam | 11 (16.7) | 1 (16.7) | 0 (0.0) | 0 (0.0) |
| Co-trimoxazole | 55 (83.3) | 3 (50.0) | 2 (100.0) | 1 (100.0) |
| Chloramphenicol | 24 (36.4) | 2 (33.3) | 0 (0.0) | 0 (0.0) |
| Clindamycin | 17 (25.8) | 1 (16.7) | 0 (0.0) | 0 (0.0) |
CoNS: Coagulase-Negative Staphylococci, VGS: Viridans Group of Streptococci
The presence of the mecA gene was confirmed with PCR, as illustrated in FIGURE 2. All the Staphylococcus spp. isolates that had the mecA gene were sensitive to vancomycin strip (M.I.C.EOxoid Limited, Wade Road, Basingstoke, and Hampshire RG24 8PW, United Kingdom). The Streptococcus pneumoniae isolated were resistant to co-trimoxazole only (100%), whereas the viridians group streptococci were resistant to both cotrimoxazole (100%) and amikacin (100%).
Discussion
This study determined the prevalence and resistant pattern of bloodstream pathogens in infants and children who presented at the emergency units of Obafemi Awolowo University Teaching Hospitals Complex, Ile-Ife, Osun State, in southwestern Nigeria.
We found the prevalence of CAB to be 27.4%. This is higher than what was obtained in similar studies in Northern (18.2%), and Central Nigeria (10.8%) 2 but lower than 48.9% obtained in another study from the South- South region of Nigeria [28]. The variation in the prevalence might be ascribed to differences in methodology or, risk factors or co-morbidity, hygiene, possibly to location because regional variation has been suggested to cause this variation [2,3,12-14,21,22,29-31].
We considered the positive blood cultures of Gram-positive bacilli, like Bacillus spp., and fungi to be contaminants since the patients had no clinical features of sepsis; the organisms could have been introduced into the culture bottle from contaminated skin at the time of collection [32]. The contamination rate of blood cultures in our study was very low at 8.2%, which is similar to a report from Ibadan4 and much lower than from another study in Iran [33].
Gram-positive bacteria were the most prevalent cultured pathogens, accounting for 79.8% of the cultured organisms. Some studies from Lagos and other developing countries have found similar higher prevalence of Gram-positive bacteria and lower prevalence of Gram-negative bacteria [34-36]. Conversely, studies from the northern and eastern Nigeria as well as Central Africa have recorded predominance of Gramnegative bacteria [3,28,29,37]. These differences could be due to hygiene, infrastructures or socioeconomic status of the people in these regions. The species of organisms isolated from the blood of the children is not much different from that reported in other studies on bacteraemia, although there are variations in the proportion as well as predominance of organisms [2,35,36,38- 41]. We found that Staphylococcus aureus was the predominant Gram-positive organisms. This is not an uncommon finding, as it has been reported in many studies; it could have been due to Staphylococcus aureus carriage on the skin, because most of the patients had soft-tissue infection initially, and some had Staphylococcus sepsis [34-37]. And also, CoNS has been significantly associated with both community and hospital acquired bacteremia [1]. Amongst the Gram-negative organisms isolated, Stenotrophomonas maltophilia predominated and this is a rather unusual finding from clinical samples of patients in this locality. Some studies done in Taiwan reported Stenotrophomonas maltophilia to be a cause of CAB [42,43].
A major finding which gives room for concern was that more than half of the isolated organisms were multi-drug resistant. Although, multi-drug resistant organisms have been reported in sub- Saharan Africa [44-47]. With could deduce from our study that antibiotic regulation is poor, this may be a contributory factor to the multidrug resistant pattern of the bacteria causing infections in our locality [48,20]. We also found out that, the commonly used drugs for empiric treatment of community acquired infections in this environment were often ineffective: Gram-negative organisms are now resistant to chloramphenicol, co-amoxiclav and ceftriaxone, which contradict what was earlier reported by Adejuyigbe et al, [20] in the same location as this study. Antibiotic regulation is poor in Nigeria, this leads to misuse of antibiotics and eventual resistance to them by bacteria. Citrobacter sedlaki and Salmonella arizonea, were both sensitive to all the tested antibiotics. The resistant pattern of these Gram-negative isolates, especially for Stenotrophomonas maltophilia, could be due to cross-resistance or the use of efflux pumps by these pathogens.
The resistance genes of the Gram-negative organism we tested for were those of ESBL genes of the 19 Gram-negative organisms, 15.8% were found to have the ESBL genes, which are fewer than reported by Kavitha, Sevitha and Sunil [35]. The only Acinetobacter haemolyticus isolated was blaCTX-M, while 50% of Hafnia alvie have both blaCTX-M and blaTEM, and 14.3% of Stenotrophomonas maltophilia have blaSHV/ blaTEM/blaCTX-M. Three types of the ESBL genes were found the locality, conforming to what was found in the North Eastern Nigeria [24]. Stenotrophomonas maltophilia is a known ESBL producing organism that has been isolated both in clinical samples and drinking water [49- 52]. In addition to multidrug-resistance ability of Acinetobacter spp., they exhibit both Extended Spectrum β-lactamase and AmpC β-lactamase resistance pattern [53-57]. Also, Hafnia alvie can acquire AmpC resistance [53,58]. Plasmid transfer could account for the presence of ESBL gene in this organisms [59,60].
Since three quarters of CAB are caused by S. aureus and CoNS with half of them resistant to cefuroxime and one third to gentamicin, these antibiotics cannot be recommended in management of CAB. Up to one-third of the Gram-positive organisms, mainly Staphylococcus aureus and CoNS were MRSA, i.e. 28.8% and 33.3% respectively had mecA genes, a rate that is similar to that reported in Mangalore by Prabhu, Bhat and Rao.35 None of the MRSA or MRCoNS bacteria isolated was found to be resistant to vancomycin. However, due to cost and nonavailability of this medication, not all the patients with MRSA infection received vancomycin therapy. Those who did not, were treated with ceftriaxone combined with gentamicin. While all the five patients treated with vancomycin recovered fully, only three (21.4%) of the 14 treated with the combination of ceftriaxone and gentamicin died, thus indicating good efficacy of this antibiotic combination. The Streptococcus pneumoniae isolated was 100% resistant to co-trimoxazole, while the Viridians Streptococci were resistant to amikacin and co-trimoxazole. Since these isolates occurred in only 3.2% of the patients co-trimoxazole and amikacin should not be included in the consideration of first line drugs for CAB in this locality.
Conclusion
The prevalence of CAB in the suspected infants and children utilizing this tertiary health facility in southwestern Nigeria was noted to be nearly 27.4%. The most common single agent isolated was Staphylococcus aureus and more than fifty percent of all the isolated pathogens were multidrug-resistant. Procalcitonin was found to be useful in excluding bacteraemia in febrile children. There is an urgent need for a review of the antibiotics in use in order to improve care. Such combinations like Piperacillin/ Tazobactam, Amikacin/Levolfoxacin or Ampicillin/Sulbactam as empirical antibiotics will probably give a good outcome in the care of these patients seen at the study location.
Acknowledgments
We thank the management of Obafemi Awolowo University Teaching Hospitals Complex, Ile-Ife, Osun State, for assistance with this study.
Source of Funding
Self-sponsored research.
Conflicts of Interest
None.
Disclosure
Data of the study was presented as an abstract poster at ASM Microbe 2017 of the American Society for Microbiology in June, 2017.
References
- Nimri LF, Batchoun R. Community-acquired bacteraemia in a rural area: predominant bacterial species and antibiotic resistance. J Med Microbiol. 53: 1045-1049 (2004).
- Obaro S, Lawson L, Essen U, et al. Community acquired bacteraemia in young children from central Nigeria-a pilot study. BMC Infect Dis. 11: 137 (2011).
- Bahwere P, Levy J, Hennart P, et al. Community-acquired bacteremia among hospitalized children in Rural Central Africa. Int J Infect Dis. 5: 180-188 (2001).
- Popoola O, Kehinde A, Ogunleye V, et al. Bacteremia among febrile patients in Nigeria. Clin Infect Dis. 69: S466-S473 (2019).
- Lambregts MMC, Bernards AT, Van Der Beek MT, et al. Time to positivity of blood cultures supports early re-evaluation of empiric broad-spectrum antimicrobial therapy. PLoS One. 14: e0208819 (2019).
- Watanabe Y, Oikawa N, Hariu M, et al. Ability of procalcitonin to diagnose bacterial infection and bacteria types compared with blood culture findings. Int J Gen Med. 9: 325-331 (2016).
- Archibald LK, Reller LB. Clinical microbiology in developing countries. Emerg Infect Dis. 7: 302-305 (2001).
- Sigauque B, Roca A, Mandomando I, et al. Community-acquired bacteraemia among children admitted to a rural hospital in mozambique. Pediatr Infect Dis J. 28: 108-113 (2009).
- Phetsouvanh R, Phongmany S, Soukaloun D, et al. Causes of community-acquired bacteraemia and patterns of antimicrobial resistance in vientiane, Laos. Am J Trop Med Hyg. 75: 978-985 (2006).
- Kumar A, Roberts D, Wood KE, et al. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med. 34: 1589-1596 (2006).
- Anthony RM, Brown TJ, French GL. Rapid diagnosis of bacteraemia by universal amplification of 23S ribosomal DNA followed by hybridization to an oligonucleotide array. J Clin Microbiol. 38: 781-788 (2000).
- Owa JA, Olusanya O. Neonatal bacteraemia in Wesley Guild hospital, Ilesa, Nigeria. Ann Trop Paediatr. 8: 80-84 (1988).
- Adejuyigbe EA, Adeodu OO, Ako-nai KA, et al. Septicaemia in high risk neonates at a teaching hospital in Ile-Ife, Nigeria. East Afr Med J. 78: 540-543 (2001).
- Calitri C, Virano S, Scolfaro C, et al. Community-acquired bloodstream infections among paediatric patients admitted to an Italian tertiary referral centre: a prospective survey. Infez Med. 20: 176-181 (2012).
- Mtove G, Amos B, Nadjm B, et al. Decreasing incidence of severe malaria and community-acquired bacteraemia among hospitalized children in Muheza, north-eastern Tanzania, 2006-2010. Malar J. 10: 320 (2011).
- Falade AG, Ayede AI. Epidemiology, aetiology and management of childhood acute community acquired pneumonia in developing countries-A review. Afr J Med Med Sci. 40: 293-308 (2011).
- Tabu C, Breiman RF, Ochieng B, et al. Differing burden and epidemiology of non-typhi salmonella bacteraemia in rural and Urban Kenya, 2006-2009. PLoS One. 7: e31237 (2012).
- Laupland KB, Gregson DB, Vanderkooi OG, et al. The changing burden of paediatric bloodstream infections in calgary, Canada 2000-2006. Pediatr Infect Dis J. 28: 114-117 (2009).
- Cisterna R, Cabezas V, Gomez E, et al. Community-acquired bacteraemia. Rev Esp Quimioter. 14: 369-382 (2001).
- Adejuyigbe EA, Ako-Nai AK, Adisa B. Bacterial isolates in the sick young infant in Ile-Ife, Nigeria. J Trop Pediatr. 50: 323-327 (2004).
- Onipede AO, Onayade AA, Elusiyan JBE, et al. Invasive bacteria isolates from children with severe infections in a Nigerian hospital. J Infect Dev Ctries. 3: 429-436 (2009).
- Clinical and Laboratory Standard Institute. Performance standards for antimicrobial susceptibility testing. Twenty fourth informational supplement update. CLSI document M100-S24U Clinical and Laboratory Standard Institute (CLSI 2014 ed.). Wayne, PA 190872016, USA. 22-180 (2014).
- Queipo-Ortuno MI, De Dios Colmenero J, Macias M, et al. Preparation of bacterial DNA template by boiling and effect of immunoglobulin G as an inhibitor in real-time PCR for serum samples from patients with brucellosis. Clin Vaccine Immunol. 15: 293-296 (2008).
- Mohammed Y, Gadzama GB, Zailani SB, et al. Characterization of extended spectrum beta-lactamase from Escherichia coli and Klebsiella species from North Eastern Nigeria. J Clin Diagn Res. 10: DC07-DC10 (2016).
- Bali EB, Acik L, Sultan N. Phenotypic and molecular characterization of SHV, TEM, CTX-M and extended-spectrum-lactamase produced by Escherichia coli, Acinobacter baumannii and Klebsiella isolates in a Turkish hospital. African J Microbio Res. 4: 650-654 (2010).
- Sajith KAK, Preetha JS, Lakshmi SY, et al. Detection of mecA genes of Methicillin-Resistant staphylococcus aureus by polymerase chain reaction. Int J Heal Rehabil Sci. 1: 64-68 (2012).
- Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 18: 268-281 (2012).
- Nwadioha S, Nwokedi E, Odimayo M, et al. Bacterial isolates in blood cultures of children with suspected septicaemia in a Nigerian Tertiary Hospital. Intern J Infec Dis. 8: 1-8 (2009).
- Meremikwu MM, Nwachukwu CE, Asuquo AE, et al. Bacterial isolates from blood cultures of children with suspected septicaemia in Calabar, Nigeria. BMC Infect Dis. 5: 110 (2005).
- Biedenbach DJ, Moet GJ, Jones RN. Occurrence and antimicrobial resistance pattern comparisons among bloodstream infection isolates from the SENTRY Antimicrobial Surveillance Program (1997-2002). Diagn Microbiol Infect Dis. 50: 59-69 (2004).
- Pfaller MA, Jones RN, Doern GV, et al. Bacterial pathogens isolated from patients with bloodstream infection: frequencies of occurrence and antimicrobial susceptibility patterns from the SENTRY antimicrobial surveillance program (United States and Canada, 1997). Antimicrob Agents Chemother. 42: 1762-1770 (1998).
- Courjon J, Demonchy E, Degand N, et al. Patients with community-acquired bacteraemia of unknown origin: clinical characteristics and usefulness of microbiological results for therapeutic issues: a single-center cohort study. Ann Clin Microbiol Antimicrob. 16: 40 (2017).
- Rahbar M, Gra-Agaji R, Hashemi S. Nosocomial blood stream infections in Imam Khomeini Hospital, Urmia, Islamic Republic of Iran, 1999-2001. East Mediterr Health J. 11: 478-484 (2005).
- Uzodimma CC, Njokanma F, Ojo O, et al. Bacterial isolates from blood cultures of children with suspected sepsis in an urban hospital in lagos: A prospective study using BACTEC blood culture system. Inter J Pedia Neonat. 16: 1-8 (2013).
- Prabhu K, Bhat S, Rao S. Bacteriologic profile and antibiogram of blood culture isolates in a pediatric care unit. J Lab Physicians. 2: 85-88 (2010).
- Isendahl J, Manjuba C, Rodrigues A, et al. Prevalence of community-acquired bacteraemia in Guinea-Bissau: an observational study. BMC Infect Dis. 14: 3859 (2014).
- Elbashier AM, Malik AG, Knot AP. Blood stream infections: microorganisms, risk factors and mortality rate in Qatif Central Hospital. Ann Saudi Med. 18: 176-180 (1998).
- Okon KO, Askira UM, Ghamba PE, et al. Childhood septicemia; retrospective analysis of bacterial pathogens and antimicrobial susceptibility pattern in maiduguri, Nigeria. New York Sci J. 7: 9-13 (2014).
- Ali J, Kebede Y. Frequency of isolation and antimicrobial susceptibility pattern of bacterial isolation from blood culture in Gondar university hospital. Ethio Med J. 46: 155-161 (2008).
- Falagas ME, Kastoris AC, Vouloumanou EK, et al. Community-acquired Stenotrophomonas maltophilia infections: a systematic review. Eur J Clin Microbiol Infect Dis. 28: 719-730 (2009).
- Gunthard H, Pennekamp A. Clinical Significance of Extra-intestinal Hafnia alvei isolates from 61 Patients and Review of the Literature. Clin Infect Dis. 22: 1040-1045 (1996).
- Chang YT, Lin CY, Chen YH, et al. Update on infections caused by Stenotrophomonas maltophilia with particular attention to resistance mechanisms and therapeutic options. Front Microbiol. 6: 893 (2015).
- Chang YT, Lin CY, Lu PL, et al. Stenotrophomonas maltophilia bloodstream infection: Comparison between community onset and hospital-acquired infections. J Microbiol Immunol Infect. 47: 28-35 (2014).
- Irenge LM, Kabego L, Kinunu FB, et al. Antimicrobial resistance of bacteria isolated from patients with bloodstream infections at a tertiary care hospital in the Democratic Republic of the Congo. S Afr Med J. 105: 752-755 (2015).
- Williams PCM, Isaacs D, Berkley JA. Antimicrobial resistance among children in sub-Saharan Africa. Lancet Infect Dis. 18: e33-e44 (2018).
- Tadesse BT, Ashley EA, Ongarello S, et al. Antimicrobial resistance in Africa: A systematic review. BMC Infect Dis. 17: 616 (2017).
- Madhi SA, Petersen K, Madhi A, et al. Increased disease burden and antibiotic resistance of bacteria causing severe community-acquired lower respiratory tract infections in human immunodeficiency virus type 1-infected children. Clin Infect Dis. 31: 170-176 (2000).
- Komolafe AO, Adegoke AA. Incidence of bacterial Septicaemia in Ile-Ife Metropolis, Nigeria. Malaysian J Micro. 4: 51-61 (2008).
- Ako-Nai AK, Oluga FA, Onipede AO, et al. The characterization of bacterial isolates from acute otitis media in Ile-Ife, Southwestern Nigeria. J Trop Pediatr. 48: 15-23 (2002).
- Adegoke AA, Stenstrom TA, Okoh AI. Stenotrophomonas maltophilia as an emerging ubiquitous pathogen: looking beyond contemporary antibiotic therapy. Front Microbiol. 8: 2276 (2017).
- Lavigne JP, Gaillard JB, Bourg G, et al. Extended-spectrum beta-lactamases-producing Stenotrophomonas maltophilia strains: CTX-M enzymes detection and virulence study. Pathol Biol. 56: 447-453 (2008).
- Adesoji AT, Ogunjobi AA. Detection of extended spectrum beta-lactamases resistance genes among bacteria isolated from selected drinking water distribution channels in Southwestern Nigeria. Biomed Res Int. 2016: 1-9 (2016).
- Ruppe E, Woerther PL, Barbier F. Mechanisms of antimicrobial resistance in Gram-negative bacilli. Ann Intensive Care. 5: 21 (2015).
- Shakibaie MR, Adeli S, Salehi MH. Antibiotic resistance patterns and extended spectrum β-lactamase production among Acinetobacter spp. Isolated from intensive care Unit of a hospital in Kerman, Iran. Antimicrob Resist Infect Control. 1: 1 (2012).
- Ibrahim ME, Mohammed A, Al-Shahrai AM, et al. Phenotypic characterization and antibiotic resistance patterns of extended spectrum β-lactamase and AmpC β-lactamase production Gram-Negative bacteria in a referral hospital, Saudi Arabia. Can J Infect Dis Med Microbiol. 2019: 1-9 (2019).
- Maravic A, Skocibusic M, Fredotovic Z, et al. Urban riverine environment is a source of multidrug-resistant and ESBL-producing clinically important Acinetobacter spp. Environ Sci Pollut Res Int. 23: 3525-3535 (2016).
- Sinha M, Srinivasa H, Macaden R. Antibiotic resistance profile & Extended Spectrum Beta-Lactamase (ESBL) production in Acinetobacter species. Indian J Med Res. 126: 63-67 (2007).
- Perez-Perez FJ, Hanson ND. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol. 40: 2153-2162 (2002).
- Kamel NA, Aboshanab KM, Abouelwafa MM, et al. Plasmid mediated extended spectrum beta-lactamase producing strains of Enterobacteriaceae isolated from diabetic foot infections in Egypt. Arch Clin Micro. 4: 1 (2013).
- Lindblom A, Sriram KK, Muller V, et al. Interspecies plasmid transfer appears rare in sequential infections with Extended-Spectrum β-Lactamase (ESBL)-producing Enterobacteriaceae. Diagn Microbiol Infect Dis. 93: 380-385 (2019).