Research Article - Pharmaceutical Bioprocessing (2017) Volume 5, Issue 2
Synthesis and x-ray crystallographic studies of azatricyclo-sulfonamide derivatives
- *Corresponding Author:
- Gunasekaran Krishnasamy
CAS in Crystallography and Biophysics
University of Madras
Guindy Campus, Chennai-600025, India
E-mail: gunaunom@gmail.com
Abstract
Azatricyclo and its derivatives occupy an important position in medicinal and pesticide chemistry with having a wide range of bioactivities such as antimicrobial, anticancer, antiinflammatory, antibacterial, antifungal and antitumor activities. Similarly, the sulfonamide groups were proved to possess various biological properties such as antimicrobial, antifungal, anticancer, anti-inflammatory, antitubercular and antiviral agents as well as HIV protease
inhibitors. In view of these biological and medicinal importances, X-ray crystallographic studies for the azatricyclo sulfonamide derivatives have been carried out. The structures were solved by direct methods and refinement was carried out using least squares procedures. The detailed information on the molecular conformation in the solid state, IUPAC name and chemical diagram of the compounds are reported.
Keywords
azatricyclo, sulfonamide, direct methods, SHELXS, bruker
Introduction
In the field of medicinal and pesticide chemistry, azatricyclo and its derivatives plays an important role by possessing various bioactivities such as antimicrobial, anticancer, anti-inflammatory, antibacterial, antifungal and antitumor activities. Similarly, the sulfonamide groups are reported to possess numerous biological properties such as antimicrobial [1,2], antifungal [3], anticancer [4,5], anti-inflammatory [6], and as well as HIV protease inhibitors [7]. Also some sulfonamides derivatives are well recognized as antimetabolite [8] and good cytotoxic against breast cancer cells [9].
Sulfonamides were the first effective chemotherapeutic agents to be utilized efficiently to prevent and cure the bacterial infection in human beings [10-13]. Recently, some sulfonamide derivatives were screened for their antioxidant activity. In addition, sulfonamides have shown carbonic anhydrase inhibition [14], COX-2 inhibition [15] and selective 5-HT receptor antagonist [16] activity. Based on the above mentioned facts, the combination of azatricyclo-sulfonamide system results in novel heterocycles which are biologically active compounds, especially as anticancer and anti-HIV agents [17]. In view of these biological and medicinal importances of the azatricyclo derivatives, X-ray crystallographic studies for the four azatricyclo derivatives have been carried out to obtain detailed information on the molecular conformation in the solid state.
Material and methods
Synthesis
N-allylated amino aldehyde were added with hydroxylamine (NH2OH.HCl) in the presence of 50% sodium hydroxide and produces the required aldoxime. On further treatment of aldoxime derivative (without isolation) with N-Chlorosuccinimide (NCS) and (Triethylamine) Et3N in the presence of carbon tetra chloride (CCl4) at room temperature over a period of 6H successfully provided the anticipated tricyclic tetrahydro isoxazoloquinoline.
Intensity data collection
X-ray diffraction intensity data were collected for all the four compounds on Bruker Kappa APEXII area-detector diffractometer [18] at the Indian Institute of Technology (IIT), Chennai equipped with graphite mono-chromated MoKα (λ=0.7103 Å) radiation and CCD detector. Crystals were cut to suitable size and mounted on a glass fiber using cyanoacrylate adhesive.
The unit cell parameters were determined from 36 frames (0.5° phi-scan) measured from three different crystallographic zones and using the technique of difference vectors. The intensity data were collected with an average fourfold redundancy per reflection and optimum resolution (0.75Ǻ). The intensity data collection, frames integration, Lp and decay correction were done using SAINT-NT (version 6.0) software. Empirical absorption correction (multi-scan) is performed using SADABS [18] program. The cell parameters and other data processing statistics of the compounds are given in the result section whereas the 2-D chemical diagram along with the IUPAC name are provided in the Supplementary File.
Results and Discussion
Structure solution and refinement
Crystal structures were solved by direct methods using SHELXS-97. The residual factors (RE) calculated based on the point atom model are 0.202, 0.210, 0.230 and 0.231, respectively for compounds I, II, III and IV. The structures were refined by the full-matrix least-squares method using SHELXL-97. All the non-hydrogen atoms were first refined isotropically and then with anisotropic displacement parameters for compound I, II, III and IV respectively. For compounds I-IV, the positions of all the hydrogen atoms were fixed by geometry and these atoms were treated by ride on their parent C atoms, with aromatic C-H distances of 0.93Å and methyl C-H distances of 0.96Å with Uiso(H)=1.5Ueq (C) for methyl H and 1.2Ueq (N,C) for other H atoms. The refinement converged to a final R-factor of 0.048 for compound I, 0.049 for compound II, 0.050 for compound III and 0.045 for compound IV (Table 1). All the four compounds have been deposited in Cambridge Structural Database (CSD) and obtained CCDC number for compound I is 1488162, compound II is 1488163, compound III is 1488176 and compound IV is 1488177.
| Parameters | I | II | III | IV |
|---|---|---|---|---|
| Empirical formula | C25H21ClN2O5S | C25H21ClN2O5S | C25H21Cl2N2O5S | C27H25N3O3S |
| Formula weight | 496.95 | 496.95 | 531.39 | 471.56 |
| Temperature | 293(2) K | 293(2) K | 293(2) K | 293(2) K |
| Wavelength | 0.71073 Å | 0.71073 Å | 0.71073 Å | 0.71073 Å |
| Crystal system | Monoclinic | Monoclinic | Monoclinic | Monoclinic |
| Space group | P21/c | P21/n | P21/c | Cc |
| Unit cell dimensions | a=11.179 (6) Å b=18.835 (1) Å c=12.137 (7) Å β=114.599 (2)º |
a=9.3900 (6) Å b=22.333 (17) Å c=11.1068 (9) Å β=99.643 (2)° |
a=17.0689 (17) Å b=11.9646 (11) Å c=24.353 (3) Å β=104.665 (3)º |
a=15.9500 (8) Å b=14.9091 (8) Å c=10.6610 (5) Å β=106.551 (2)° |
| Volume | 2323.7 (2) Å3 | 2296.4 (3)Å3 | 4811.5 (8) Å3 | 1986.1 (3)Å3 |
| Z, Calculated density | 4, 1.421 mg/m3 | 4, 1.437 mg/m3 | 8, 1.467 mg/m3 | 4, 1.289 mg/m3 |
| Absorption coefficient | 0.295 mm-1 | 0.298 mm-1 | 0.397 mm-1 | 0.167 mm-1 |
| F(000) | 1032 | 1032 | 2192 | 992 |
| Crystal size (mm) | 0.25 × 0.20 × 0.20 | 0.20 × 0.25 × 0.20 | 0.25 × 0.20 × 0.20 | 0.20 × 0.25 × 0.20 |
| θ range for data collection | 2 to 30.48º | 1.82 to 27.93º | 1.23 to 24.15º | 1.23 to 24.15º |
| Limiting indices | -15 ≤ h ≤ 15 -26 ≤ k ≤ 26 -16 ≤ l ≤ 17 |
-12 ≤ h ≤ 11 -29 ≤ k ≤ 29 -14 ≤ l ≤ 14 |
-19 ≤ h ≤ 19 -13 ≤ k ≤ 13 -26 ≤ l ≤ 27 |
-23 ≤ h ≤ 23 -21 ≤ k ≤ 21 -15 ≤ l ≤ 13 |
| Reflections collected/unique |
48042/6492 [Rint=0.038] |
30812/5514 [Rint=0.068] |
38647/7635 [Rint=0.048] |
24506/7062 [Rint=0.029] |
| Completeness to theta | 100% | 100% | 99.4% | 93.0% |
| Refinement method | Full-matrix least-squares on F2 | Full-matrix least-squares on F2 | Full-matrix least-squares on F2 | Full-matrix least-squares on F2 |
| Data/restraints/parameters | 6492/0/307 | 5514/0/307 | 7635/0/631 | 7062/2/307 |
| Goodness-of-fit on F2 | 0.917 | 0.776 | 1.01 | 0.997 |
| Final R indices [I>2σ(I)] | R1=0.0481 | R1=0.0497 | R1=0.0508 | R1=0.0454 |
| R indices (all data) | wR2=0.1152 R1=0.0943 wR2=0.1489 |
wR2=0.1448 R1=0.1248 wR2=0.2190 |
wR2=0.1320 R1=0.0910 wR2=0.1684 |
wR2=0.1010 R1=0.0904 wR2=0.1240 |
| Largest diff. peak and hole | 0.304 and -0.370 e.Å-3 | 0.376 and -0.454 e.Å-3 | 0.284 and -0.334 e.Å-3 | 0.218 and -0.300 e.Å-3 |
Table 1. Crystal data for compounds I, II, III and IV.
Crystal structure of compound I
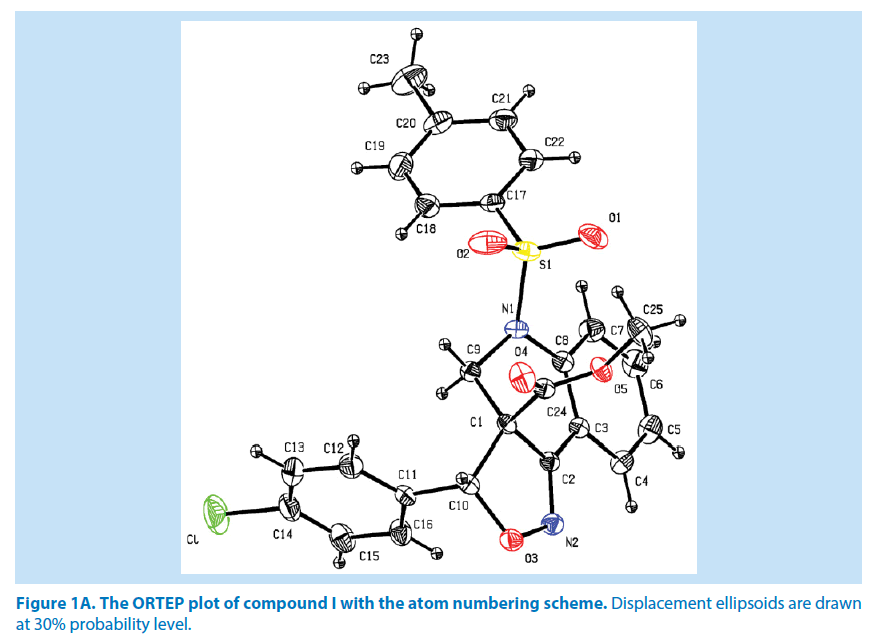
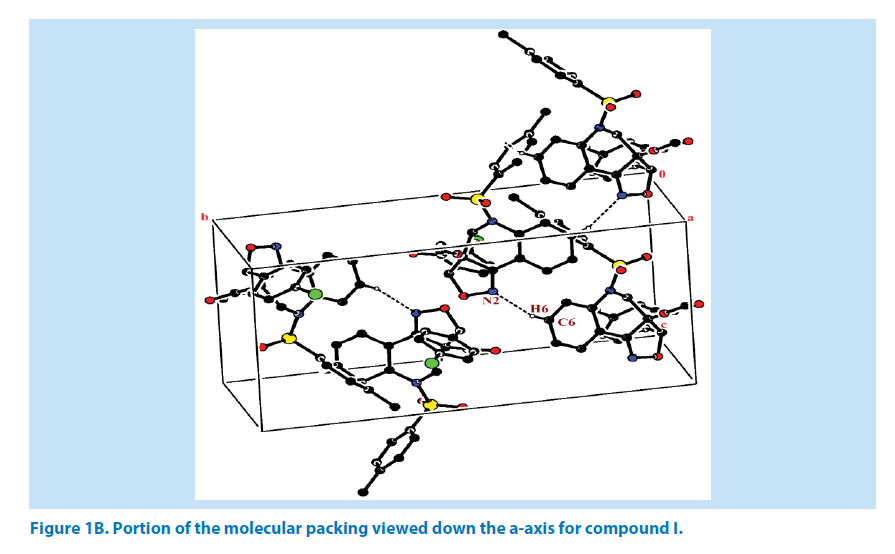
In compound I, the azatricyclo ring (C1- C9/C10/N1/N2/O3) is essentially in planar system expect for the atoms C1 and C9 which are deviated with maximum root mean square (rms) deviation of 0.337 (1) Å and -0.366 (1) Å respectively. The azatricyclo ring are almost perpendicular to the choloro phenyl ring (C11-C16, CL) which forms the dihedral angles of 86.87 (1)°. The bond length of the atom S1- O1, S1-O2, S1-C17, and S1-N1 distances are 1.422 (2) Å, 1.427 (2) Å, 1.761 (2) Å and 1.656 (2) Å respectively. These values are comparable with similar structures [19,20]. As a result of the electron-withdrawing character of the phenylsulfonyl group, the N-C sp2 bond lengths, viz. N1-C8 [1.430 (3) Å] and N1-C9 [1.476 (3) Å] are longer than the mean value of 1.355 (1) Å reported for N atoms with planar configurations [21]. It has been observed that S atom exhibits significant deviation from that of a regular tetrahedron, with the largest deviation being O-S-O [O1-S1-O2=120.46 (1)°] whereas O-S-N deviates by angle [O1-S1-N1=108.96 (1)°]. The widening of the angles may be due to repulsive interactions between the two short S=O bonds, similar to what is observed in related structures [22]. The sum of bond angles around N1 is 357.76 (2)° which indicate that the pyridine ring is in accordance with sp2 hybridization [23]. In the azatricyclic ring, six membered pyridine ring (C1-C3/C8/N1/C9) adopts a sofa conformation while five membered oxazole ring (C1/C2/N2/ O3/C10) adopts an envelope conformation with respect to its puckering parameters [24] for pyridine ring as q2=0.386 (4) Å, Φ2=-47.35 (1)° and for oxazole as q2=0.251 (1) Å, Φ2=147.01 (1)°. It can be further confirmed from the leastsquare planes analysis that the atom C9 of pyridine and C1 of oxazole ring are found to be deviated by maximum deviation of -0.366 (1) Ǻ and 0.377(1) Ǻ respectively. Crystal packing of the compound is stabilized by C6-H6…N2 inter molecular hydrogen bond interactions (Figures 1A and 1B; Table 2).
| Compounds | D-H…A | D-H | H…A | D…A | D-H…A |
|---|---|---|---|---|---|
| I | C6-H6…N2i | 0.930(1) | 2.492(1) | 3.379(1) | 159 |
| II | C6-H6…O3 i | 0930(1) | 2.472(1) | 3.365(1) | 157 |
| III | C19A-H19A…O4Ai | 0.930(1) | 2.580(1) | 3.505(4) | 173 |
| C19B-H19B…O4Bii | 0.930(1) | 2.530(1) | 3.446(3) | 169 | |
| IV | C10-H10…O2i | 0.980(1) | 2.332(1) | 3.228(1) | 152 |
| C15-H15…O1iii | 0.930(2) | 2.515(1) | 3.346(1) | 149 |
Table 2. Hydrogen bond interactions for compounds I-IV [Å and º].
Crystal structure of compound II
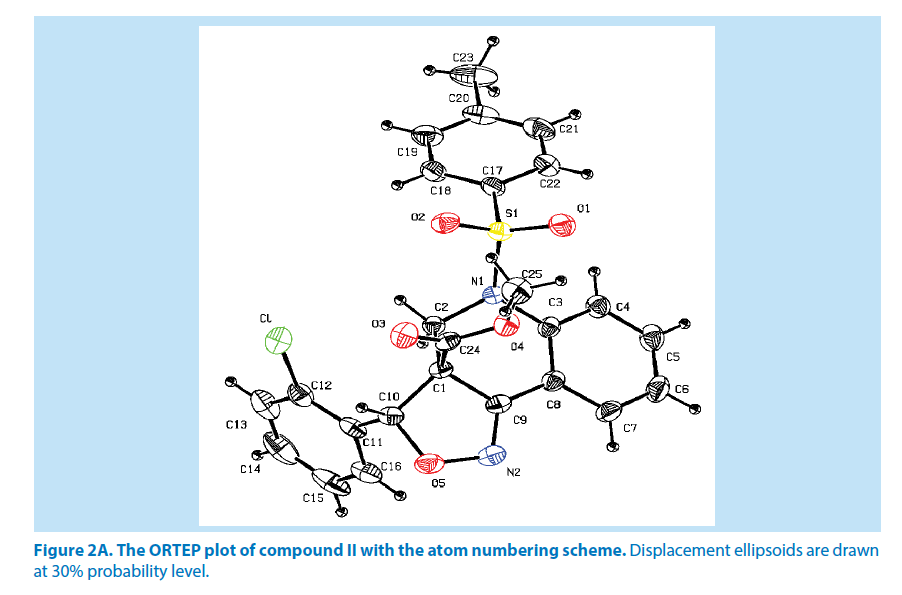
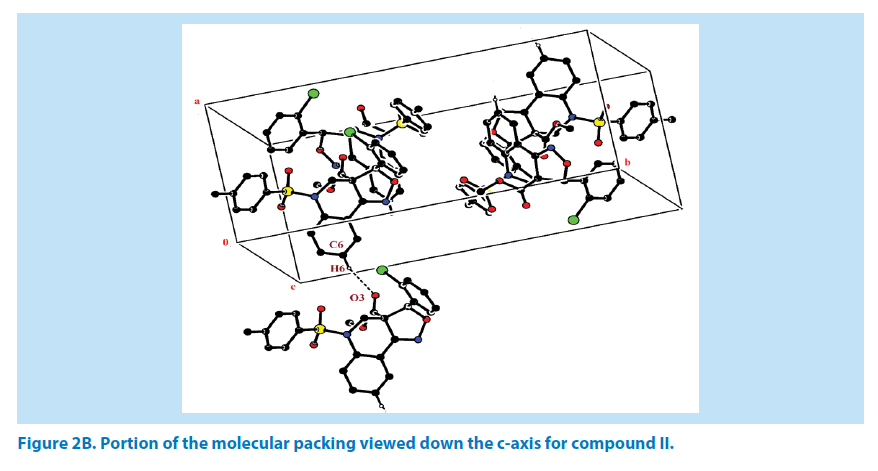
In compound II, the azatricyclo ring (C1-C9/ C10/N1/N2/O5) system is essentially planar expect the atoms C1 and C2 are deviated with a maximum rms deviation of -0.357 (1) Å and 0.310 (1) Å respectively. The azatricyclo ring are almost perpendicular to the chloro phenyl ring (C11-C16, CL) which forms the dihedral angles of 86.09 (1)°. The bond length of the atom S1-O1, S1-O2, S1-C17, and S1-N1 distances are 1.425 (2) Å, 1.426 (2) Å, 1.759 (3) Å and 1.650 (2) Å respectively. These values are comparable with the previously reported structures [20]. As a result of the electron-withdrawing character of the phenylsulfonyl group, the N-C sp2 bond lengths, viz. N1-C2 [1.468 (4) Å] and N1-C3 [1.435 (4) Å] are longer than the mean value of 1.355 (1) Å reported for N atoms with planar configurations [21]. As observed in compound I, the S atom in compound II also exhibits significant deviation from that of a regular tetrahedron, with the largest deviation being O-S-O [O1-S1-O2=120.13 (2)°] while O-S-N [O1-S1-N1] is deviated by an angle of 106.29 (1)°. The widening of the angles may be due to repulsive interactions between the two short S=O bonds, similar to what is observed in related structures [22]. The sum of bond angles around N1 is 356.55 (2)° which indicates that the pyridine ring is in accordance with sp2 hybridization [23]. As for the azatricyclic ring of compound II, the six membered pyridine ring (C1/C2/ N1/C3/C8/C9) and five membered oxazole ring (C1/C9/N2/O5/C10) adopts sofa and envelop conformation respectively with respect to its puckering parameters [24] for pyridine ring as q2=0.342 (3) Å, Φ2=54.53 (1)° and for oxazole as q2=0.274 (6) Å, Φ2=-34.14 (1)°. The least-square planes analysis further confirms that C2 atom of pyridine and C1 atom of oxazole ring are found to be deviated by maximum deviation of 0.310 (1) Ǻ and -0.357 (1) Ǻ respectively. Crystal packing of the compound is stabilized by C6-H6…O3 inter molecular hydrogen bond interactions (Figures 2A and 2B; Table 2).
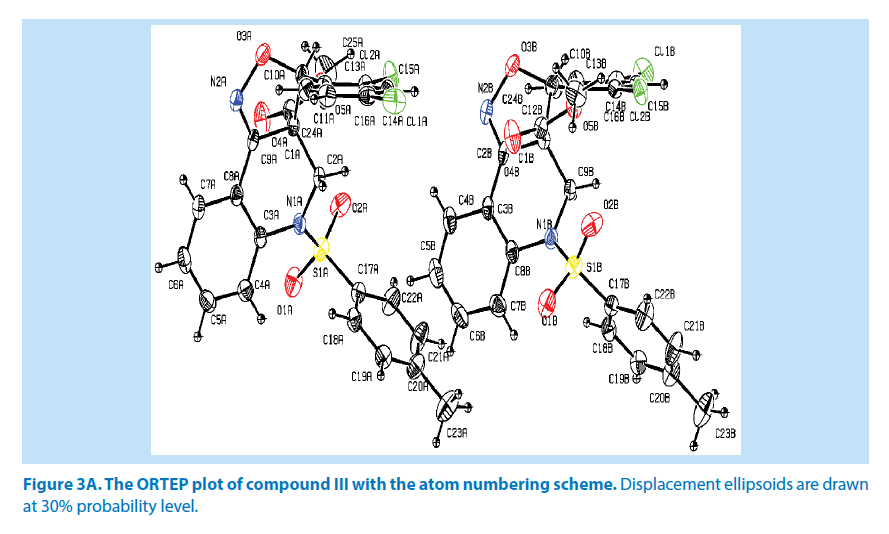
Crystal structure of compound III
In the title compound III, it consists of two molecules (A and B) in the asymmetric unit. Molecule A of azatricyclo ring (C1A-C9A/C10A/ N1A/N2A/O3A) system is essentially planar expect the atoms C1A and C2A are deviated with a maximum rms deviation of 0.338 (1) Å and -0.377 (1) Å respectively. Similarly molecule B of azatricyclic ring, the atom C1B and C9B are deviated with a maximum rms deviation of 0.338 (1) Å and -0.368 (1) Å respectively. The azatricyclo ring of molecule (A &B) are almost perpendicular to the dichloro phenyl ring which forms the dihedral angles of 81.72 (1)° for molecule A and 75.50 (1)° for molecule B. In case of the bond length of atoms, distances between S1A-O1A, S1A-O2A, S1A-C17A and S1A-N1A in molecule A are 1.423 (3) Å, 1.431 (3) Å, 1.756 (4) Å and 1.648 (3) Å respectively while the distances between S1B-O1B, S1B-O2B, S1B-C17B and S1B-N1B in molecule B are 1.427 (3) Å, 1.427 (3) Å, 1.746 (4) Å and 1.651 (3) Å respectively. These values are comparable with the previously reported structures [20]. As a result of the electron-withdrawing character of the phenylsulfonyl group, the N-C sp2 bond lengths, viz. N1A-C2A [1.479 (5) Å], N1AC3A [1.432 (5) Å], N1B-C9B [1.479 (4) Å] and N1B-C8B [1.431 (5) Å] are longer than the mean value of 1.355 (1) Å reported for N atoms with planar configurations [21]. The S atom also exhibits significant deviation from that of a regular tetrahedron, with the largest deviation being O-S-O where [O1A-S1A-O2A] of molecule A is around 118.6 (2)° and [O1BS1B- O2B] of molecule B is around 118.2 (2)°.
As observed in the compounds I and II, O-S-N is deviated by angles [O1A-S1A-N1A] with 108.04 (1)° for molecule A and [O1B-S1B-N1B] with 107.89 (2)° for molecule B. The widening of the angles may be due to repulsive interactions between the two short S=O bonds, similar to what is observed in related structures [22]. The sum of bond angles around N1A is 359.3 (1)° and N1B is 359.1 (1)° which indicate that the molecule (A&B) of pyridine ring is in accordance with sp2 hybridization [23].
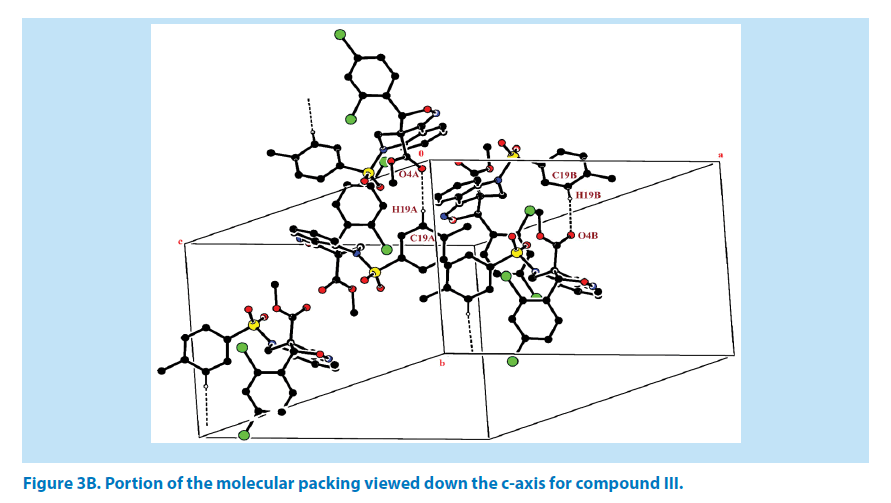
For the azatricyclo ring in molecule (A&B), six membered pyridine ring of molecule A (C1A/ C2A/N1A/C3A/C8A/C9A) and molecule B (C1B/C2B/N1B/C3B/C8B/C9B ) adopts sofa conformation whereas five membered oxazole ring of molecule A (C1A/C2A/N2A/O3A/ C10A) and molecule B (C1B/C9B/N2B/O5B/ C10B) adopts envelop conformation with respect to its puckering parameters [24] as follows. For the pyridine ring of molecule A, q2=0.278 (3) Å, Φ2=146.89 (1)° and molecule B q2=0.381 (1) Å, Φ2=128.95 (1)° whereas for the oxazole of molecule A, q2=0.278 (3) Å, Φ2=146.89 (1)° and molecule B q2=0.278 (1) Å, Φ2=69.64 (1)°. Crystal packing of the compound is stabilized by C19A-H19A…O4A and C19B-H19B…O4B inter molecular hydrogen bond interactions (Figures 3A and 3B; Table 2).
Crystal structure of compound IV
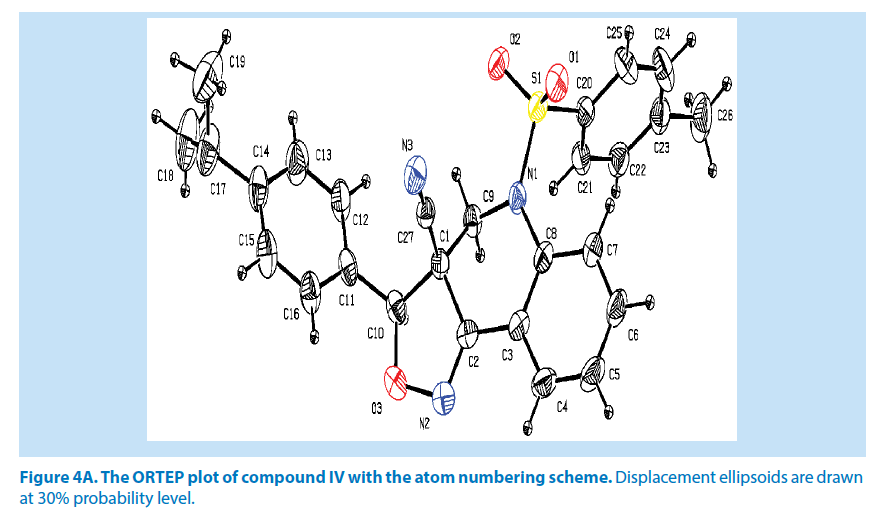
In compound IV, the azatricyclo ring (C1-C9/ C10/N1/N2/O3) system is essentially planar except the atoms C8 and C10 are deviated with maximum rms deviation of -0.368 (1) Å and 0.338 (1) Å respectively. The azatricyclo ring are almost perpendicular to the methyl phenyl ring (C20-C25) which forms the dihedral angles of 87.74 (1)° similarly on other side of dimethyl phenyl (C11-C16) ring tilted with respect to the azatricyclic ring which forms the dihedral angle of 36.46 (1)°. The bond length of the atom S1- O1, S1-O2, S1-C20, and S1-N1 distances are 1.414 (2) Å, 1.427 (1) Å, 1.753 (3) Å and 1.647 (2) Å. These values are comparable with similar structures. As a result of the electron-withdrawing character of the phenyl sulfonyl group, the N-C sp2 bond lengths, viz. N1-C8 [1.429 (3) Å] and N1-C9 [1.463 (3) Å] are longer than the mean value of 1.355 (1) Å reported for N atoms with planar configurations [21].
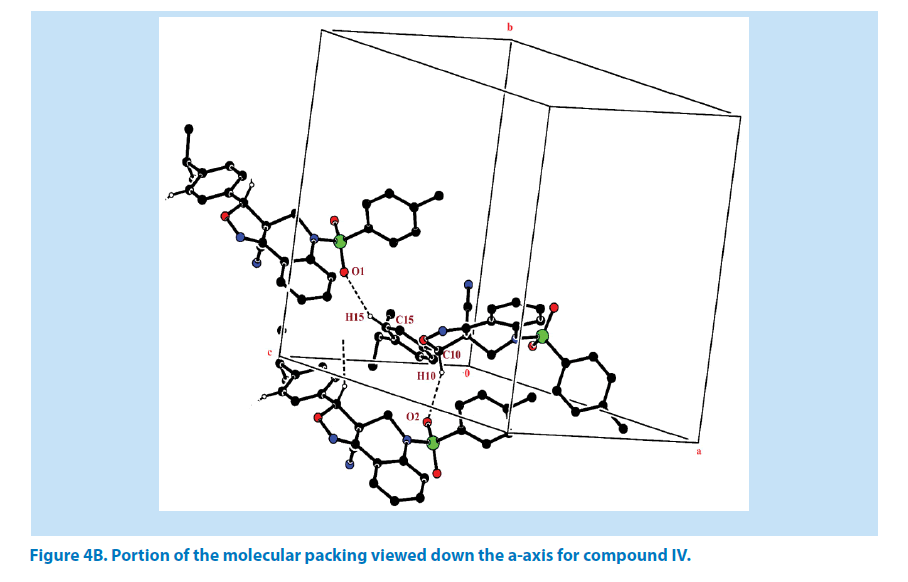
In addition to this, it was observed that S atom exhibits significant deviation from that of a regular tetrahedron, with the largest deviation being O-S-O [O1-S1-O2=119.44 (2)°] and O-S-N deviates by angles [O1-S1-N1=108.57 (1)°]. The widening of the angles may be due to repulsive interactions between the two short S=O bonds, similar to what is observed in related structures [22]. The sum of bond angles around N1 is 359.89 (1)° which indicate that the pyridine ring is in accordance with sp2 hybridization [23]. In the azatricyclic ring, six membered pyridine ring (C1-C3/C8/N1/C9) adopts sofa conformation and five membered oxazole ring (C1/C2/N2/ O3/C10) adopts envelop conformation with respect to its puckering parameters (24) for pyridine ring as q2=0.369 (3) Å, Φ2=-38.65 (1)° and for oxazole as q2=0.238 (8) Å, Φ2=143.53 (1)°. From the least-square planes analysis it can be confirmed further that the atom C8 of pyridine and C10 of oxazole ring are deviated by maximum deviation of -0.368 (1) Å and 0.338 (1) Å respectively. Crystal packing of the compound is stabilized by C10-H10…O2 and C15-H15…O1 inter molecular hydrogen bond interactions (Figures 4A and 4B; Table 2).
Conclusion
The azatricyclo derivatives were crystallized and structure determination was carried out using X-ray diffraction. All the crystals were diffracted well and intensity statistics found agreeable with crystallographic standards. The structures were solved by direct methods using SHELXS and refinement was carried out using (SHELXL) least squares procedures. All the four compounds were deposited in the Cambridge Structural Database (CSD). In future, all these compounds can play a role in aiding Structure Based Drug Design (SBDD) where their inhibitory activity will be carried out through in silico as well as in vivo studies against the suitable protein targets. Further optimizations will be employed to increase their inhibitory potency and be utilized in combating many dreadful diseases.
Acknowledgement
LK acknowledges UGC, India for funding in the form of UGC-BSR Research Fellowship in Sciences.
References
- Genç Y, Özkanca R, Bekdemir Y. Antimicrobial activity of some sulfonamide derivatives on clinical isolates of Staphylococus aureus. Ann. Clin. Microbiol. Antimicrob. 7(1), 17 (2008).
- Özbek N, Katırcıoğlu H, Karacan N et al. Synthesis, characterization and antimicrobial activity of new aliphatic sulfonamide. Bioorg. Med. Chem. 15(15), 5105–5109 (2007).
- Briganti F, Scozzafava A, Supuran CT. (1997) Sulf onylamido derivatives of aminoglutethimide and their copper (II) complexes: a novel class of antifungal compounds. Eur. J. Med. Chem. 32(11), 901–910 (1997).
- Mun J, Jabbar AA, Devi NS et al. Design and in Vitro Activities of N–Alkyl–N–[(8–R–2, 2–dimethyl–2 H–chromen–6–yl) methyl] heteroarylsulfonamides, Novel, Small–Molecule Hypoxia Inducible Factor–1 Pathway Inhibitors and Anticancer Agents. J. Med. Chem. 55(15), 6738–6750 (2012).
- El–Sayed NS, El–Bendary ER, El–Ashry SM et al. Synthesis and antitumor activity of new sulfonamide derivatives of thiadiazolo [3, 2–a] pyrimidines. Eur. J. Med. Chem. 46(9), 3714–3720 (2011).
- Borne RF, Peden RL, Waters IW et al. Anti‐inflammatory activity of para‐substituted N‐benzenesulfonyl derivatives of anthranilic acid. J. Pharm. Sci. 63(4), 615–617 (1974).
- Clercq E. New developments in anti–HIV chemotherapy. Curr. Med. Chem. 8(13), 1543–1572 (2001).
- Mengelers MJB, Hougee PE, Janssen LHM et al. Structure‐activity relationships between antibacterial activities and physicochemical properties of sulfonamides. J. Vet. Pharm. Ther. 20(4), 276–283 (1997).
- Mirian M, Zarghi A, Sadeghi S et al. Synthesis and cytotoxic evaluation of some novel sulfonamidederivativesagainst a few human cancer cells. Iran. J. Pharm. Res. 10(4), 741–748 (2011).
- Bhusare SR, Pawar RP, Vibhute YB. Synthesis and antibacterial activity of some new 2–(substituted phenyl sulfonamido)–6–substituted benzothiazoles. Ind. J. Heterocyclic. Chem. 11(1), 79–80 (2001).
- Bhusari KP, Khedekar PB, Umathe SN et al. Synthesis of 8–bromo–9–substituted–1, 3–benzothiazolo–[5, 1–b]–1, 3, 4–triazoles and their anthelmintic activity. Ind. J. Heterocyclic. Chem. 9(4), 275–278 (2000).
- Ahmed B, Khan SA, Alam T. Synthesis and antihepatotoxic activity of some heterocyclic compounds containing the 1, 4–dioxane ring system. Int. J. Pharm. Sci. 58(3), 173–176 (2003).
- Shekar BC, Roy K, De AU. (2001). Synthesis of some new p–toluene sulfonamido glutaramides. Ind. J. Heterocyclic. Chem. 10(3), 237–238 (2001).
- Supuran CT, Ilies MA, Scozzafava A. Carbonic–anhydrase inhibitors – part 29 – interaction of isozyme–i, isozyme–ii and isozyme–iv with benzolamide–like derivatives. Eur. J. Med. Chem. 33(9), 739–751.
- Dannhardt G, Fiebich BL, Schweppenhäuser J. COX–1/COX–2 inhibitors based on the methanone moiety. Eur. J. Med. Chem. 37(2), 147–161 (2002).
- Bromidge SM, Clarke SE, King FD et al. Bicyclic piperazinylbenzenesulphonamides are potent and selective 5–HT 6 receptor antagonists. Bioorg. Med. Chem. Lett. 12(10), 1357–1360 (2002).
- Tabaković K, Tabaković I, Trkovnik M et al. Chemistry of Coumarins.‐Nucleophilic Substitutions of 4‐Chloro‐3‐nitrocoumarin with Hard and Soft Nucleophiles. Eur. J. Org. Chem. 1983(11), 1901–1909 (1983).
- Bruker. APEX2, SAINT and SADABS. Bruker AXS Inc. Madison, Wisconsin, USA (2010).
- Sheldrick GM. A short history of SHELX. Acta. Crystall. Sec. A. 64(1), 112–122 (2008).
- Ranjith S, Subbiahpandi A, Namitharan K et al. 2–(4–Chlorophenyl)–1, 5–diphenyl–3–tosylimidazolidin–4–one. Acta. Crystal. Sec.67(4), o843–o843 (2011).
- Allen FH, Kennard O, Watson DG et al. Tables of bond lengths determined by X–ray and neutron diffraction. Part 1. Bond lengths in organic compounds. J. Chem. Soc. 2(12), S1–S19 (1987).
- Kavitha T, Thenmozhi M, Dhayalan V et al. 8, 9–Dimethoxy–5–phenylsulfonyl–5H–benzo [b] carbazole. Acta. Crystallographica. Sec. 66(5), o1071–o1071 (2010).
- Beddoes RL, Dalton L, Joule JA et al. The geometry at nitrogen in N–phenylsulphonyl–pyrroles and–indoles. The geometry of sulphonamides. J. Chem. Soc. 2, (6), 787–797 (1986).
- Cremer DT, Pople JA. General definition of ring puckering coordinates. J. Am. Chem. Soc. 97(6), 1354–1358 (1975).