Mini Review - Interventional Cardiology (2021) Volume 13, Issue 3
The association of Paraoxonase 1 (PON1) gene polymorphisms with the coronary artery disease
- Corresponding Author:
- Sana Ashiq
Centre for Applied Molecular Biology,
University of the Punjab,
Lahore,
Pakistan
E-mail: sanaashiq72@gmail.com
Received date: April 16, 2021; Accepted date: April 30, 2021; Published date: May 07, 2021
Abstract
Non-Communicable Diseases (NCDs) are the major reason for death in today’s world. Among these non-communicable diseases, the prevalence of Cardiovascular Diseases (CVDs) ranked highest. Worldwide among these CVDs Coronary Artery Disease (CAD) is the most common cause of mortality and morbidity. It is a multifactorial disorder and is influenced by many genes to environment and gene to gene interactions. The lack of physical activity, unhealthy diet routine, stress and smoking are the most common lifestyle-related risk factors. The family history, ethnicity and genetic polymorphisms including the Single Nucleotide Polymorphisms (SNPs) in genes also increase the susceptibility to the CAD. One of the most important polymorphisms is in the gene paraoxonase 1 (PON1). Single nucleotide polymorphism in this gene can alter its hydrolytic activity as the mutant allele carriers are more susceptible to CAD as the wild allele type is more effective in inhibiting the oxidation of Low-Density Lipoprotein Cholesterol (LDLC) as compared to the mutant isoform. Thus, the purpose of this review is to summarize the association between the polymorphisms in the PON1 gene and coronary artery disease. Moreover, we also conclude that the several in-silico tools and pharmacogenomics approaches can play a potential role in the treatment of coronary artery diseases.
Keywords
PON1 • Polymorphisms • Coronary artery diseases
Introduction
Cardiovascular Diseases (CVDs) belong to the category of disorders that mainly consist of cardiac and the blood circulatory system. Coronary Artery Disease (CAD) is one of the most prevalent forms of cardiovascular disorders [1]. Lack of oxygen supply to the heart and vasoconstriction are the two main factors that are responsible to instigate CAD as the balance between oxygen supply and its demand is highly disturbed [2]. Around the globe, the mortality rate of CVDs is found to be nearly 31% and it is estimated that almost 7.4 million expiries are occurred due to CAD alone. In addition, the prevalence rate of CAD is ranked highest among developing countries. CAD patients’ can be presented either with acute or chronic symptoms, i.e. stable angina, cardiac failure and acute coronary syndrome [3]. One of the key biomarkers that plays a significant role in atherosclerosis is the Paraoxonase 1 (PON1) gene. It is bound to High-Density Lipoprotein Cholesterol (HDLC) surface, hence shows a key role in antioxidant activity of HDL [4]. The oxidation of Low-Density Lipoprotein Cholesterol (LDLC) is quite critical to avoid the risk of CAD. In this regard, the PON1 gene plays a significant role as it averts the oxidation of LDLC by hydrolyzing the lipid peroxides. Single Nucleotide Polymorphisms (SNPs) in the PON1 gene resulted in the mutation that ultimately disturbs the catalytic activity of the gene [5]. To date, several SNPs were studied in the PON1 gene but two polymorphisms in the coding region were extensively studied. One is Q192R (rs662) and the other is the L55M (rs854560) polymorphism in which a single nucleotide substitution can affect the activity of the gene, hence showed a significant association with coronary artery disease [4]. Also, many previous studies provide evidence of CAD association with polymorphism of the PON1 gene among the various ethnic groups [6-8].
The purpose of the current review is to critically analyze the relationship of polymorphisms in the paraoxonase 1 gene with CAD that provides us the more definite results. In addition, we have also revealed the current pharmacogenomics approaches to CAD treatment.
Risk Factors
The Framingham Heart Study (FHS) was published in 1957 and acknowledged as the first scientific literature that presented the conception of “CAD associated risk factors”. This study established the epidemiologic associations of blood pressure, cigarette smoking, and cholesterol levels to the occurrence of CAD. The outcomes of the study were groundbreaking as it reformed the medicine approach practiced at that time. The FHS findings helped not only to identify the CAD risk factors but also shown risk factors that can be controlled or that cannot be controlled. The controlled risk factors are also called modifiable and included diabetes, raise blood pressure, obesity, lack of physical activity, stress, smoking, unhealthy dietary routine and hypercholesterolemia. The conventional risk factors are those that cannot be modified or controlled, i.e., race, sex, age and family history [9].
The clinical phenotype of CVDs can be determined by knowing interactions between environmental and genetic factors. Many Genome‐Wide Association Studies (GWAS) proved that CAD is strongly associated with a wide range of genetic variants. Moreover, current research reports that ethnic variations among populations contribute to the genetic variation in CAD risk. Lifestyle is a leading factor that contributes in the pathogenesis of the cardiovascular diseases [10].
Association of Paraoxonase 1 Gene Polymorphisms with Coronary Artery Disease
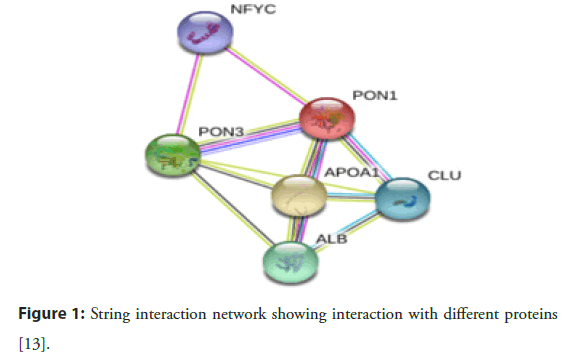
Abraham Mazur was the scientist who pointed out that there is an animal enzyme that can detoxify the organophosphates. In the early 1950s, this enzyme was discovered and named “Paraoxonase” due to its ability to hydrolyze an important toxic metabolite of pesticide parathion called paraoxon. Over the past few years, paraoxonase is accepted as an important enzyme family and applied in many types of research due to its antiatherogenic action and capability to detoxify. In 1996, Primo-Parmo with his coworkers discovered that the paraoxonase gene family is a multigene comprised of PON1, PON2 and PON3. Many investigations verified that PON1, PON2 and PON3 genes have almost 65% amino acid and 70% nucleotide level similarity and are situated alongside in the long arm of chromosome 7 q21.3-22.1 in humans. Despite the similarities, PON1, PON2 and PON3 demonstrate variances in terms of their substrate specificity, intracellular positioning and catalytic actions. Although all the paraoxonase family possesses hydrolytic activity but the research is more documented on the PON1 due to it protective role against the LDL and HDL oxidation, hence play a vital role in the inhibition of atherosclerosis and other diseases associated with oxidative stress [11]. Furthermore, it is associated with apolipoprotein 1 (APOA1) which stabiles the structure of the gene. Thus, due to any Single Nucleotide Polymorphism (SNP), the stability of the gene structure can be affected which ultimately affects the hydrolytic activity of the PON1 [12]. These SNPs may progress toward the disease by modulating its effect on various biochemical and anthropometric parameters [1]. Figure 1 explains the role of PON1 in CAD pathogenesis [13].
Figure 1: String interaction network showing interaction with different proteins [13].
To date, a total of two hundred single nucleotide polymorphisms have been reported in the PON1 gene including the seven SNPs in the promoter region, five SNPs in the exonic region, one hundred and seventy-one in the intronic region and fifteen polymorphisms in 3’-untranslated regions. The functional SNPs include both the promoter and coding regions of the gene. The two well- documented polymorphisms include the L55M and Q192R, in which the risk allele affects the hydrolytic activity and it is less stable in inhibiting the oxidation of LDL as compared to the wild allele. However, many studies revealed the variation in the results as American Caucasians revealed the R allele as a risk allele for the CAD as compared to the findings reported from the Korean, Spanish and British Caucasians population [2] (Table 1).
| Author | SNP | Findings | Reference |
|---|---|---|---|
| Shabana et al. (2018) | Q192R | p-value 0.02 | [1] |
| Kaur et al. (2018) | Q192R L55M |
p-value 0.0001 p-value 0.326 |
[4] |
| Munshi et al. (2018) | Q192R L55M |
p-value <0.0001 p-value >0.05 |
[8] |
| Rad et al. (2020) | Q192R | p-value >0.05 | [14] |
Table 1: Summarizes the main findings from several recent studies.
Pharmacogenomics: Current Status and Future Perspective
The contribution of genetics to interindividual variability in drug response is the core part of pharmacogenomics. Genetically based differences in drug metabolism now gain more importance due to their wider clinical applications and development toward the personalized medicines. As the single nucleotide polymorphisms may affect the efficacy of the cardiovascular drugs thus based on this genetic information it can help in designing the cardiovascular drugs and can also guide the clinician about the selection of the drug that can optimize the therapeutic benefit [15]. The different in-silico tools can further help to check the gene interaction with various ligands that can ultimately help in drug designing [16]. Sirtuin 1 is the vital regulator for the inflammation mechanisms. Moreover, study results also suggest the association between Sirtuin 1 and PON1 as for the Sirtuin 1 gene expression there is a need for normal PON1 gene activity [17]. Sirtuin 1 repression is associated with the progression of chronic diseases as it inactivates the various drug metabolism. In addition, proteomic analyses are important to determine the severity and treatment of CAD [18].
Many study findings also suggest an association among the PON1 and ABCB1 gene variants has been implicated as modulating clopidogrel activity. The major limitation of these genetic studies is the smaller sample size thus large population size studies are required which may provide us better information that how to implement genomic medicine in the electronic medical record context. In addition, genetic risk scores can reclassify the individuals thus provide us the better treatment options [19].
Conclusion
We conclude that the PON1 genetic polymorphism plays a crucial role in coronary artery diseases. This PON1 polymorphism may progress toward the CAD by modulating its effect on several anthropometric and biochemical parameters. Finally, we suggest that the use of various in-silico tools, pharmacogenomics approaches and genetic risk scores can help in better identification and treatment of coronary artery disease.
References
- Shabana NA, Ashiq S, Ijaz A, et al. Genetic risk score (GRS) constructed from polymorphisms in the PON1, IL-6, ITGB3, and ALDH2 genes is associated with the risk of coronary artery disease in Pakistani subjects. Lipids Health Dis. 17(1): 1-7 (2018).
- Ashiq S, Ashiq K, Shabana S, et al. Prevalence and role of different risk factors with emphasis on genetics in development of pathophysiology of coronary artery disease (cad). Pakistan Heart Journal. 52(4): (2019).
- Ashiq S, Ashiq K. The Role of paraoxonase 1 (PON1) gene polymorphisms in coronary artery disease: A systematic review and meta-analysis. Biochem Genet. 18: 1-21 (2021).
- Kaur S, Bhatti GK, Vijayvergiya R, et al. Paraoxonase 1 gene polymorphisms (Q192R and L55M) are associated with coronary artery disease susceptibility in Asian Indians. Int J Diabetes Metab. 21(1-4): 38-47 (2018).
- Moreno-Godínez ME, Galarce-Sosa C, Cahua-Pablo JÁ, et al. Genotypes of common polymorphisms in the PON1 gene associated with paraoxonase activity as cardiovascular risk factor. Arch Med Res. 49(7): 486-96 (2018).
- Hernández-Díaz Y, Tovilla-Zárate CA, Juárez-Rojop IE, et al. Effects of paraoxonase 1 gene polymorphisms on heart diseases: Systematic review and meta-analysis of 64 case-control studies. Medicine. 95(44): e5298 (2016).
- Zeng Q, Zeng J. A meta-analysis on relationship between paraoxonase 1 polymorphisms and atherosclerotic cardiovascular diseases. Life Sciences. 232: 116646 (2019).
- Munshi R, Panchal F, Chaurasia A, et al. Association between Paraoxonase 1 (PON1) gene polymorphisms and PON1 enzyme activity in Indian patients with Coronary Artery Disease (CAD). Curr Pharmacogenomics Person Med (Formerly Current Pharmacogenomics). 16(3): 219-29 (2018).
- Hajar R. Risk factors for coronary artery disease: historical perspectives. Heart Views. 18(3): 109 (2017).
- Malakar AK, Choudhury D, Halder B, et al. A review on coronary artery disease, its risk factors, and therapeutics. J Cell Physiol. 234(10): 16812-23 (2019).
- Alim Z, Kilic D, Koksal Z, et al. Assessment of the inhibitory effects and molecular docking of some sulfonamides on human serum paraoxonase 1. J Biochem Mol Toxicol. 31(10): e21950 (2017).
- Paul S, Solayman M, Saha M. In silico analysis of the functional and structural impacts of non-synonymous single nucleotide polymorphisms in the human paraxonase 1 gene. Int J Bioautomation. 19(3): 275 (2015).
- PON1 Gene. Gene Cards. (2021).
- Rad NM, Sardabi EH. Evaluation of the role of paraoxonase1 Q192R polymorphism in coronary artery disease. Turkiye Klinikleri J Cardiovasc Sci. 32(1): 9-13 (2020).
- Anderson JL, Carlquist JF, Home BD, et al. Cardiovascular pharmacogenomics: Current status, future prospects. J Cardiovasc Pharmacol Ther. 8(1): 71-83 (2003).
- Ashiq S, Ashiq K. In-silico analysis of PON1, PON2 and PON3 genes role in coronary artery disease. IJCBDD. 13(4): 415-23 (2020).
- Breitenstein A, Wyss CA, Spescha RD, et al. Peripheral blood monocyte Sirt1 expression is reduced in patients with coronary artery disease. PLoS One. 8(1): e53106 (2013).
- Martins IJ. Sirtuin 1, a diagnostic protein marker and its relevance to chronic disease and therapeutic drug interventions. EC Pharmacol Toxicol. 6(4): 209-15 (2018).
- Roden DM. Cardiovascular pharmacogenomics: Current status and future directions. J Hum Genet. 61(1): 79-85 (2016).