Review Article - Neuropsychiatry (2017) Volume 7, Issue 3
The N-methyl-D-aspartate (NMDA) receptor in skin, dorsal root ganglion and spinal cord: an ideal target gene for RNA interference therapy for pain relief
- *Corresponding Author:
- P.H. Tan
Department of Anesthesiology
E-DA Hospital, I-Shou University
Kaohsiung, Taiwan
Tel: +886-7-6150011, ext. 2532
Fax: +886-7-3423348
E-mail: tanphphd@hotmail.com
Abstract
The NMDA receptor plays an important role in induction of pain hyperalgesia and allodynia by tissue trauma. Although the analgesic effect of NMDA receptor antagonists has been well reported in various experimental study and clinical pain trial, serious side effects were reported and limited their use in clinical medication. To obtain NMDA receptor antagonist with less side effects, an alternative approach is to use subtype-selective NMDA receptor antagonists. However, the effects of current available NMDA receptor antagonist’s drug are not absolute specific for antagonizing specific NMDA receptor subtypes.
Keywords
NMDA receptor, Antinociception, Spinal cord, Skin, Dorsal root ganglion
Introduction
RNA interference (RNAi) is an evolutionarily conserved process leading to post-transcriptional gene silencing medicated by short doublestranded RNA (dsRNA) molecules. Discovered in 1998 [1], the RNAi pathway, the one of the most important technological breakthroughs in modern biology, allowing us to study protein function and validate the expression of specific gene in mammalian system. Until now it has become a valuable research tool for gene knockdown studies, pathway analysis, phenotype analysis, and drug target discovery. The development of pain is associated with complex interaction of neurotransmitters and proteins. RNAi has become a promising tool to study the expression of gene and protein involved in pain processing pathways and to treat inflammatory and neuropathic pain [2-5]. In the past years, RNAi have already been applied in many pain studies, and microRNAs (miRNAs) are a rising field in pain research and treatment. Short dsRNA can be generated by exogenous or endogenous short hairpin RNA and then exerts breakage of mRNA. The 3’ untranslated region (UTR) of a single gene is predicted to be targeted by several different miRNAs with bioinformatical analysis [6]. miRNAs might be a promising tool to manage inflammatory and neuropathic pain by modulating the expression of gene and protein involved in pain processing pathways. There is accumulating evidence to implicate the importance of N-methyl-D-aspartate (NMDA) receptors to the induction and maintenance of central sensitization during pain states. Recently, RNAi was applied to silence the NMDA receptor in spinal cord, dorsal root ganglion and skin for pain relief due to its high specificity. In this review, we will review the pain studies applying gene knockdown of NMDA receptor subunits in skin, dorsal root ganglion and spinal cord by RNAi.
Mechanism of RNA Interference
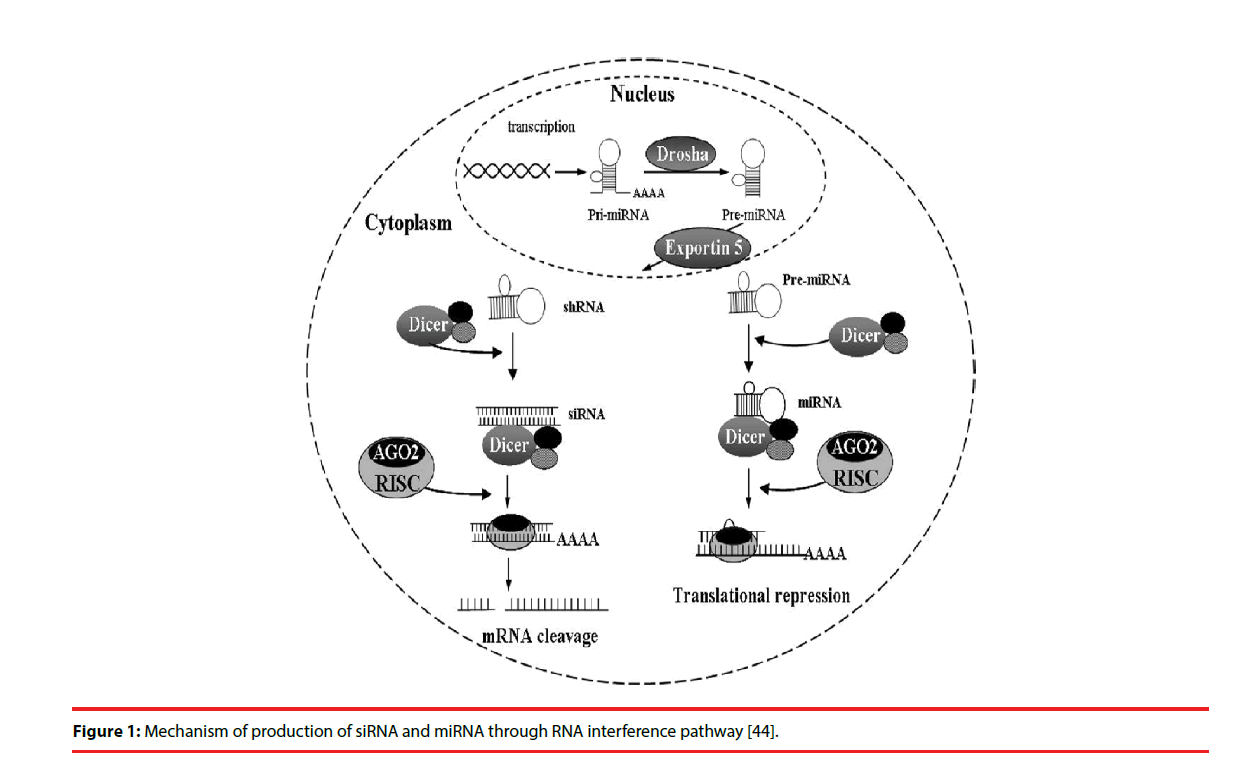
Two types of small RNA molecules – small interfering RNAs (siRNAs) and miRNAs- are known to mediate RNAi. miRNAs are small noncoding RNAs of 18-25 nucleotides that regulate gene expression. The RNAi is initiated by the RNase III enzyme Dicer, which cleaves long dsRNA molecules of exogenous or endogenous origin [7,8] into short double-stranded fragments of 21 to 23-nucleotide siRNAs. These siRNAs are processed to single guide (antisense) strands by selective cleavage of the passenger (sense) strand by the enzyme Argonaute 2 [9,10]. The antisense strands were then incorporated into the RNA-induced silencing complex (RISC) [11], which then identified a complementary sequence in mRNA and induced cleavage by Argonaut 2 in the RISC [12]. Subsequently, these cleaved products are targeted for degradation, which results in reduction of target protein levels.
In most eukaryotic cells, miRNAs are mostly transcribed as primary transcripts of long primary miRNAs (pri-miRNA) from intragenic or intergenic regions by RNA polymerase II (Pol II) [13,14]. The primary transcripts are further processed by RNase III enzyme Drosha in the nucleus into a hairpin intermediate of ~ 65 nucleotides, called pre-miRNA [15]. The pre-miRNA is transported to cytoplasm by the nuclear export factor Exportin-5 [16]. The pre-miRNAs are then processed by another ribonuclease called Dicer into a mature doublestranded miRNA of about 18-25 nucleotides in the cytoplasm [17]. The guide strand or mature miRNA is incorporated into the RISC, which separates the miRNA duplex as single strand with sequence complementary to specific target mRNA [17]. Once programmed, the RISC can down-regulate the expression of homologous mRNAs (Figure 1).
Figure 1: Mechanism of production of siRNA and miRNA through RNA interference pathway [44].
Subunit Composition and Localization of the NMDA Receptor
Spinal glutamate receptors play an important role in the pain transmission and facilitation. For example, α-amino-3-hydroxy-5-methyl- 4-isoazolepropionate (AMPA) and kainate receptors mediate monosynaptic reflexes [18] and acute nociceptive responses [19]. Conversely, NMDA receptors are implicated in polysynaptic spinal pathways [20] and in chronic nociceptive responses. Intrathecal administration of NMDA has been shown to induce pain hyperalgesia in mice and rats [21]. In addition, following various forms of peripheral nociceptive stimulation increased spinal glutamate release was observed [22]. Three types of genes encoding NMDA receptor subunits have been identified, namely NMDA-R1 (NR1), A–D isoforms of NMDA-R2 (NR2A–D), and NMDA-R3 subunit [23]. Functional NMDA-R2B receptor subunit protein (NR2B)-containing NMDA receptors are formed by their coassembly with the obligatory NR1 subunits [24]. While NR1 distributes ubiquitously in the central nervous system (CNS), NR2 subunits exhibit regional distribution and predominate in forebrain structures [25]. In a recent study, NR2B protein was found in the spinal cord, especially located in the superficial dorsal horn [26]. Although the efficacy of NMDA receptor antagonists in various experimental and clinical pain situations has been well documented, their use as analgesics is limited by serious side effects such as memory impairment, psychotomimetic effects, ataxia, and motor incoordination. To use specific subtype NMDA receptor antagonism is a promising way to obtain effective NMDA receptor antagonist analgesics and avoid side effects. However, the effect of current available pharmacological NMDA receptor antagonists is not absolutely selective for specific NMDA receptor subtypes. Therefore, siRNAs which are able to inactive NMDAR subunit gene expression are used.
Gene Knockdown of NMDA Receptor Subunits in Spinal Cord
Temporal and spatial gene deletion in the CNS can be achieved by the Cre–loxP recombination system [27,28]. This is a way of deleting an essential DNA sequence between the two loxP sites catalyzed by Cre introduced by Tsien et al. [27]. South, et al. modified the technique by employing a recombinant adeno-associated virus (rAAV) encoding a green fluorescent protein/ Cre recombinase (GFP/Cre) fusion protein developed by Kaspar, et al. [28] to deliver Cre recombinase to the lumbar spinal cord dorsal horn (SCDH) of adult floxed NR1 mice, allowing the specific deletion of SCDH NMDA receptors NR1 subunit [29]. They reported that expressions of NR1 subunit mRNA and NMDA currents but not AMPA currents were reduced more than 80% in lamina II neurons. The heat or cold paw-withdrawal latencies, mechanical threshold, or motor function were not altered by NR1 knock-out. However, formalin-induced pain in the paw was reduced by 70%. Their results indicate that the postsynaptic NR1 receptor subunit in the lumbar SCDH play an essential role in inducing central sensitization and pain transmission stimulated by peripheral noxious input.
Elbashir and his colleagues reported RNAi can be elicited effectively by well-defined 21-base duplex siRNAs [7]. Tan, et al. [3] used siRNAs that inhibit NR2B gene expression to evaluate the effect of gene silencing on antinociception in a rat model of formalin-induced pain. The results showed that gene silence of NR2B subunit by siRNA decreased the expression of NR2B mRNA and protein and further abolished formalin-induced pain behaviors. The peak inhibition of NR2B mRNA and protein occurred on day 3 and day 7 following intrathecal injection of 5 μg of siRNA-NR2B. These data showed that investigation of functional gene expression in the whole animal behavior model for the pain management by intrathecal siRNAs is feasible. Similar results were reported after intrathecal injection of short hairpin RNA (shRNA) targeting NR2B [30]. After that, Garraway, et al. [4] reported in vivo spatial knockdown of the expression and function of the NR1 subunit in spinal cord by rAAV plasmid encoded- NR1 siRNAs. They found that 60% to 75% knockdown of NR1 mRNA and protein was found in the rAAV plasmid injected area. A single injection of the vector resulted in spatial knockdown for at least 6 months. The vector-derived knockdown of NR1 expression in SCDH could prevent mechanical allodynia at 24 and 48 h after injection of complete Freund’s adjuvant (CFA) into the hindpaws of rats but could not affect acute thermal or mechanical withdrawal thresholds of paw. Garraway and his colleagues [5] also investigated the effects of intraparenchymal administration of rAAV-encoded NR1 siRNA sequence into the SCDH on formalin-induced nociception in adult rats. They found that vector-derived siRNAs effectively produced spatial knockdown of NR1 gene expression that effectively attenuated formalin-induced pain behaviors and NR1 phosphorylation in the animals. These evidence proved that intrathecal siRNA targeting NMDA receptor can attenuate inflammatory pain and is a powerful tool in the investigation of spinal cord functional gene expression in the context of whole animal behavior.
Gene Knockdown of NMDA Receptor Subunits in Skin and Dorsal Root Ganglion
NMDA receptors are ubiquitously expressed in sensory neurons and axons. In the lumbar dorsal root ganglion (DRG), virtually all cells express the NR1 subunit [31]. In the periphery, Coggeshall and collegues showed that approximately 50% of the myelinated and 20-30% of the unmyelinated axons in plantar (mixed motor and sensory) and sural nerves immunostained for NR1 [32]. In addition, the NMDA receptors also existed on postganglionic sympathetic efferents, Schwann cells, and keratinocytes in the skin and affected peripheral nociceptive sensitization [33]. During inflammation, the number of ionotropic glutamate receptors, particularly NR1 in sensory axons increases, which contributes to peripheral sensitization in inflammation [34]. Thus, silencing of the gene that encodes for the NMDA receptor NR1 subunit in skin and DRGs can produce an antinociceptive effect. To study the antinociceptive effects of siRNA-mediated gene silencing, Tan, et al. first investigated the effect of knockdown of NMDA receptor NR1 subunit in skin by NR1 synthetic siRNA in rat models of formalin- and CFA-induced pain [35]. The behavioral data and analysis of skin tissue showed that 1 nmol NR1 siRNA administered intradermally effectively diminished the level of NR1 mRNA and protein in skin and attenuated the formalin and CFA-induced nociception. The administration of NR1 siRNA produced antinociceptive effect and inhibited NR1 expression for about 7 days. Furthermore, the fluorescence intensities of NR1-positive neurons and mRNA of NR1 in L5 DRGs ipsilateral to a NR1 siRNA-injected hind paw were significantly lower than those in the contralateral L5 DRG. This implies that the siRNA injected intradermally will be transported in a retrograde fashion to the ipsilateral DRG, subsequently silencing the expression of NR1. This finding was consistent with that reported by Thakor, et al. who showed that plasmid DNA or a short hairpin RNA injected subcutaneously appeared to undergo retrograde transport to L4 and L5 DRGs [36]. This study suggest that NR1 siRNA administered subcutaneously is an effective way in treating inflammatory pain induced or maintained by peripheral nociceptor activity and support the potential application of siRNA in studying peripheral nociceptive processes. We previously examined whether administration of a vector encoding NR1 shRNA could provide a prolonged antinociceptive effect in a rat model of formalin-induced pain [37]. We found that subcutaneously administering 10 μg of the NR1 shRNA-encoded vector resulted in a significant decrease in the mRNA and protein levels of NR1 on the 7th and 14th day after administration. A partial recovery of the mRNA and protein levels was found on the 21st day and a full recovery of the mRNA and protein levels of NR1 was noted on the 28th day after administration of 10 μg NR1 shRNA. This suggests that subcutaneously administering NR1 shRNA could provide prolonged pain relief that is induced or maintained by peripheral nociceptor activity.
Viral vectors are commonly used in vivo and most vectors use polymerase (Pol) III promoters such as H1 and U6 or the minimal CMV promoter to drive the expression of siRNA [38]. However, excessive expression of siRNA driven by Pol III promoters may result in off-target disruption of gene expression and may induce cellular toxicity [39]. Studies have shown that shRNAs targeting specific genes of interest can be inserted into a pre-miRNA backbone, such as miR30. Vector delivery of such shRNA-embedded miRNAs results in long chain transcripts containing naturally occurring miRNA and molecules involved in the RNAi pathway that are produced by the endogenous miRNA processing mechanism [40,41]. The gene knockdown effect of a miRNA-based shRNA whose expression is driven by Pol II promoters is 12 times more potent than that of traditional shRNA whose expression is driven by Pol III promoters. Tan, et al. examined whether subcutaneously injecting a lentiviral vector encoding a NR1 miR30-embedded shRNA (shRNAmir) could produce more efficient gene silencing effect and antinociception on formalin-and CFA-induced pain in rats [42]. Tan, et al. found that intradermal injection of 1, 5, and 10 μg of shRNAmir significantly inhibited flinch responses and that administration of 5 μg of shRNAmir resulted in the attenuation of CFA-induced mechanical allodynia, but did not affect the time spent on the rotarod. There was also a significant reduction in the percentage of NR1- and pERK-positive neurons in the DRG ipsilateral to shRNAmir-treated paws. NR1 knockdown in skin and the DRG did not alter the basal nociceptive threshold or the NMDA receptor composition in the DRG. Administration of 5 μg of shRNAmir resulted in more than an 80% decrease in both NR1 mRNA and protein expression in skin. Suppression of NR1 mRNA and protein expression was also noted after 3 days at a dose of 1 μg but was most significant at a dose of 5 μg, reflecting a fast onset of expression at very low doses of the CMV-shRNAmir system in this study. Potential adverse consequences of RNAi include activation of the interferon response and off-target gene silencing effects. In this study, the level of interferon-α mRNA expression did not increase significantly and the expression of NR2 subunits did not significantly change after administration of NR1 shRNAmir, indicating that the system did not elicit an interferon response or have an off-target effect. The results of this study demonstrate that peripheral administration of a vector-encoded NR1 shRNAmir is a promising therapy for persistent inflammatory pain.
Conclusion
The contribution of the NMDA receptor in tissue damage-induced pain hypersensitivity has been well established [43]. RNAi is often more potent and longer lasting than other nucleic acid-based approaches and, compared to gene targeting by homologous recombination, is less expensive and time consuming and not limited to a particular mammalian species [4]. Recent studies show that siRNAs or vectors encoding shRNAs that target NMDA receptors located in the central or peripheral nervous systems can prevent injury-induced pain hypersensitivity, suggesting that siRNA-based therapeutics are promising strategies for treating injury-induced pain. Recent studies have also shown that miRNAs are potential diagnostic markers or therapeutic targets in various pathophysiological conditions, especially in cancer and cardiovascular diseases. RNA interference is an emerging but powerful genetic tool with great potential for aiding in the development of future therapeutic drugs for the treatment of pain. However, for translation of RNAi therapeutics to clinic, several problems must be conquered such as effective delivery system and activation of immune system. Besides this, challenges with respect to the use of miRNAbased therapeutics such as poor cellular uptake or a low bioavailability as well as off-target effects or long-term safety concerns in humans remain to be further explored. Therefore, to maximize siRNAs and miRNAs potency and minimize off target toxicity and immunogenicity are most important for the future prospects of RNAi-based therapy [44].
Acknowledgements
This work was supported by Taiwan Ministry of Science and Technology Grant NSC 103-2314-B-214-003-MY3, 105-2314-B-214- 002-MY3 and E-Da Hospital Grants EDPJ 103054, 103055, 104068, EDAHP 104028 and NCKUEDA 10503.
References
- Fire A, Xu S, Montgomery MK, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391(6669), 806-811 (1998).
- Dorn G, Patel S, Wotherspoon G, et al. siRNA relieves chronic neuropathic pain. Nucleic. Acids. Res 32:e49 (2004).
- Tan PH, Yang LC, Shih HC, et al. Gene knockdown with intrathecal siRNA of NMDA receptor NR2B subunit reduces formalin-induced nociception in the rat. Gene. Ther 12(1), 59-66 (2005).
- Garraway SM, Xu Q, Inturrisi CE. Design and evaluation of small interfering RNAs that target expression of the N-methyl-D-aspartate receptor NR1 subunit gene in the spinal cord dorsal horn. J. Pharmacol. Exp. Ther 322(3), 982-988 (2007).
- Garraway SM, Xu Q, Inturrisi CE. siRNA-mediated knockdown of the NR1 subunit gene of the NMDA receptor attenuates formalin-induced pain behaviors in adult rats. J. Pain 10(4), 380-90 (2009).
- Krek A, Grün D, Poy MN, et al. Combinatorial microRNA target predictions. Nat. Genet 37(5), 495-500 (2005).
- Elbashir SM, Harborth J, Lendeckel W, et al. Duplexes of 21-nucleotide RNAs mediates RNA interference in cultured mammalian cells. Nature 411(6836), 494-498 (2001).
- Bernstein E, Caudy AA, Hammond SM, et al. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409(6818), 363-366 (2001).
- Matranga C, Tomari Y, Shin C, et al. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell 123(4), 607-620 (2005).
- Rand TA, Petersen S, Du F, et al. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell 123(4), 621-629 (2005).
- Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes. Dev 15(2), 188-200 (2001).
- Hammond SM, Boettcher S, Caudy AA, et al. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293(5532), 1146-1150 (2001).
- Lee Y, Jeon K, Lee JT, et al. MicroRNA maturation: stepwise processing and subcellular localization. EMBO. J 21(17), 4663-70 (2002).
- Cullen BR. Transcription and processing of human microRNA precursors. Mol. Cell 16(6), 861-865 (2004).
- Lee Y, Ahn C, Han J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 425(6956), 415-419 (2003).
- Yi R, Qin Y, Macara IG, et al. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes. Dev 17(24), 3011-3016 (2003).
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2), 281-297 (2004).
- Long SK, Smith DA, Siarey RJ, et al. Effect of 6-cyano-2,3-dihydroxy-7-nitro-quinoxaline (CNQX) on dorsal root-, NMDA-, kainate- and quisqualate-mediated depolarization of rat motoneurones in vitro. Br. J. Pharmacol 100(4), 850-854 (1990).
- Dickenson AH, Sullivan AF. Evidence for a role of the NMDA receptor in the frequency dependent potentiation of deep rat dorsal horn nociceptive neurones following C fibre stimulation. Neuropharmacology 26(8), 1235-1238 (1987).
- Watkins JC, Evans RH. Excitatory amino acid transmitters. Annu. Rev. Pharmacol. Toxicol 21165-21204 (1981).
- Meller ST, Dykstra C, Gebhart GF. Production of endogenous nitric oxide and activation of soluble guanylate cyclase are required for N-methyl-D-aspartate-produced facilitation of the nociceptive tail-flick reflex. Eur. J. Pharmacol 214(1), 93-96 (1992).
- Sorkin LS, Westlund KN, Sluka KA, et al. Neural changes in acute arthritis in monkeys. IV. Time-course of amino acid release into the lumbar dorsal horn. Brain. Res. Rev 17(1), 39-50 (1992).
- Kutsuwada T, Kashiwabuchi N, Mori H, et al. Molecular diversity of the NMDA receptor channel. Nature 358(6381), 36-41 (1992).
- Masuko T, Kashiwagi K, Kuno T, et al. A regulatory domain (R1-R2) in the amino terminus of the N-methyl-D-aspartate receptor: effects of spermine, protons, and ifenprodil, and structural similarity to bacterial leucine/isoleucine/valine binding protein. Mol. Pharmacol 55(6), 957-969 (1999).
- Monyer H, Burnashev N, Laurie DJ, et al. Developmental and regional expression in the rat brain and functional properties of four NMDA receptors. Neuron 12(3), 529-40 (1994).
- Boyce S, Wyatt A, Webb JK, et al. Selective NMDA NR2B antagonists induce antinociception without motor dysfunction: correlation with restricted localisation of NR2B subunit in dorsal horn. Neuropharmacology 38(5), 611-623 (1999).
- Tsien JZ, Huerta PT, Tonegawa S. The essential role of hippocampal CA1 NMDA receptor-dependent synaptic plasticity in spatial memory. Cell 87(7), 1327-1338 (1996).
- Kaspar BK, Vissel B, Bengoechea T, et al. Adeno-associated virus effectively mediates conditional gene modification in the brain. Proc. Natl. Acad. Sci. U S A 99(4), 2320-2325 (2002).
- South SM, Kohno T, Kaspar BK, et al. A conditional deletion of the NR1 subunit of the NMDA receptor in adult spinal cord dorsal horn reduces NMDA currents and injury-induced pain. J. Neurosci 23(12), 5031-5040 (2003).
- Zhang RX, Yan XB, Gu YH, et al. Gene silencing of NR2B-containing NMDA receptor by intrathecal injection of short hairpin RNA reduces formalin-induced nociception in C57BL/6 mouse. Int. J. Neurosci 123(9), 650-656 (2013).
- Sato K, Kiyama H, Park HT, et al. AMPA, KA and NMDA receptors are expressed in the rat DRG neurones. Neuroreport 4(11), 1263-1265 (1993).
- Coggeshall RE, Carlton SM. Ultrastructural analysis of NMDA, AMPA, and kainate receptors on unmyelinated and myelinated axons in the periphery. J. Comp. Neurol 391(1), 78-86 (1998).
- Carlton SM. Peripheral excitatory amino acids. Curr. Opin. Pharmacol 1(1), 52-56 (2001).
- Du J, Zhou S, Coggeshall RE, et al. N-methyl-D-aspartate-induced excitation and sensitization of normal and inflamed nociceptors. Neuroscience 118(2), 547-562 (2003).
- Tan PH, Chia YY, Chow LH, et al. Gene knockdown of the N-methyl-D-aspartate receptor NR1 subunit with subcutaneous small interfering RNA reduces inflammation-induced nociception in rats. Anesthesiology 112(6), 1482-1493 (2010).
- Thakor D, Spigelman I, Tabata Y, et al. Subcutaneous peripheral injection of cationized gelatin/DNA polyplexes as a platform for non-viral gene transfer to sensory neurons. Mol. Ther 15(12), 2124-2131 (2007).
- Tan PH, Yu SW, Lin VC, et al. RNA interference-mediated gene silence of the NR1 subunit of the NMDA receptor by subcutaneous injection of vector-encoding short hairpin RNA reduces formalin-induced nociception in the rat. Pain 152(3), 573-581 (2011).
- Xia H, Mao Q, Paulson HL, et al. siRNA-mediated gene silencing in vitro and in vivo. Nat. Biotechnol 20(10), 1006-1010 (2002).
- Grimm D, Streetz KL, Jopling CL, et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature 441(7092), 537-541 (2006).
- Zeng Y, Cai X, Cullen BR. Use of RNA polymerase II to transcribe artificial microRNAs. Methods. Enzymol 392371-392380 (2005).
- Silva JM, Li MZ, Chang K, et al. Second-generation shRNA libraries covering the mouse and human genomes. Nat. Genet 37(11), 1281-1288 (2005).
- Liu CC, Cheng JT, Hung KC, et al. Lentiviral vector-encoded microRNA-based shRNA-mediated gene knockdown of NMDA receptors in skin reduces pain. Brain. Behav 7(1), e00587 (2017).
- Woolf CJ, Thompson SW. The induction and maintenance of central sensitization is dependent on N-methyl-D-aspartic acid receptor activation; implications for the treatment of post-injury pain hypersensitivity states. Pain 44(3), 293-299 (1991).
- Tan PH, Yanh LC, Ji RR. Therapeutic potential of RNA interference in pain medicine. Open. Pain. J 2(1), 57-63 (2009).