Research Article - International Journal of Clinical Rheumatology (2023) Volume 18, Issue 9
The Relationship of Disease Activity and Colchicine Resistance in Patients with Familial Mediterranean Fever with CYP3A4, CYP2D6, and MDR1 Gene Variants
Busra Bilim Turkcan1*, Aslı Toylu2, Elif Comak 3, Mustafa Koyun3 , Sema Akman3
1Department of Pediatrics, Akdeniz University School of Medicine, Antalya, Turkey
2Department of Medical Genetics, Akdeniz University School of Medicine, Antalya, Turkey
3Department of Pediatric Nephrology, Akdeniz University School of Medicine, Antalya, Turkey
Department of Pediatrics, Akdeniz University School of Medicine, Antalya, Turkey
E-mail: busra_bilim@hotmail.com
Received: 27-Aug-2023, Manuscript No. fmijcr-23-111380; Editor assigned: 29- Aug-2023, Pre-QC No. fmijcr-23-111380 (PQ); Reviewed: 12-Sep-2023, QC No. fmijcr-23-111380; Revised: 14-Sep-2023, Manuscript No. fmijcr-23-111380 (R); Published: 21-Sep-2023, DOI: 10.37532/1758- 4272.2023.18(9).160-167
Abstract
The Relationship of Disease Activity and Colchicine Resistance in Patients with Familial Mediterranean Fever with CYP3A4, CYP2D6, and MDR1 Gene Variants ABSTRACT Background: Colchicine is the mainstay of Familial Mediterranean Fever (FMF) treatment. The molecules responsible for colchicine metabolism are CYP3A4, CYP2D6, and MDR1. We aimed to determine the frequency of MDR1 c.3435C>T, CYP3A41b c.-392G>A, CYP2D6*4 c.1934G>A, and CYP2D6*3c.2637Adel variants in healthy and patient groups, and to examine the relation between these variants and colchicine resistance. Methods: This cross-sectional study was performed between January 2019 and Fabruary 2019. The patient group consisted of children aged 3-18 years having a mutation in the MEFV gene who were diagnosed according to Turkish Pediatric FMF criteria. Children without any underlying chronic disease were considered as the control group. Data were obtained from face-to-face appointments and medical records. Results: Overall, 124 children with FMF and 60 healthy children were enrolled. The variant distributions were similar in both groups. There was no statistically significant difference between the variant distribution and colchicine resistance (p>0.05). However, the dose of colchicine used was significantly lower in those with the MDR1 mutant allele and those with CYP2D6*4 mutation compared to other patients (p<0.05). Conclusion: Although several allele variants responsible for colchicine metabolism are unrelated to colchicine resistance, MDR1 and CYP2D6*4 mutations can predict a lower dose of colchicine need.
Keywords
Familial Mediterranean fever • Colchicine resistance • Genetic variation
Introduction
Familial Mediterranean Fever (FMF) is an autosomal recessive disorder linked to mutations in the MEFV gene. It is characterized by recurrent fever and episodes of inflammation involving the peritoneum, pleura, joints, and/or skin resulting from impaired function of the “pyrin/marenostrin” protein, a product of the MEFV gene [1,2].
Colchicine, taken orally, is absorbed from the jejunum and ileum. It is metabolized in the liver; the majority is excreted with the stool, and 10-20% is excreted with the kidneys [4]. CYP3A4 and CYP2D6 (from the CYP450 family of enzymes) and MDR1, a transport protein, are molecules responsible for the metabolism of colchicine. Individuals are divided into slow, medium, fast, and ultrafast metabolizers according to the genotypic difference in these molecules [5]. Since the elimination of the drug is reduced in those with slow and moderate metabolism, the drugrelated side effects are increased. However, those with ultrafast metabolizers are risky in terms of the frequency of side effects as a result of therapeutic failure and the rapid formation of the active drug [6].
There are significant differences between the recommended doses of colchicine for pediatric patients. It was recently suggested that individual differences in colchicine treatment response and frequency of side effects may be related to colchicine metabolizing enzyme variants [3]. There are limited number of studies evaluating the importance of CYP3A41B, CYP2D6, and MDR1 genetic variants, which are responsible for colchicine metabolism, in FMF and their effects on colchicine metabolism, and the findings are controversial [7-9]. Considering the challenges of biological treatment options, predicting the colchicine resistance earlier is highly important. Since there are controversial findings, and there is a lack of data in pediatric age, we aimed to evaluate the relationship between MDR1 c.3435C>T, CYP3A41b c.-392G>A, CYP2D6*4 c.1934G>A and CYP2D6*3 c.2637Adel variants distribution and disease severity, colchicine response and colchicine dose requirements in children with FMF.
Material and Method
Study group and data collection:
In our clinic, 124 children aged 3-18 years having a mutation in the MEFV gene who were diagnosed according to Turkish Pediatric FMF criteria were included in the study.
Inclusion criteria:
All the patients were on colchicine therapy, and all had been followed up for at least 12 months, with a minimum three outpatient clinic visits. The healthy group was also included in the study, and it was requested to show the distribution of the selected variants in the general population. The control group consisted of 60 healthy volunteer children who had no family history of FMF and symptoms suggestive of FMF, whose physical examination findings and examinations did not reveal any signs of chronic inflammation, and who were matched in age and gender.
All the children included in the study and their families were given information and voluntary consent form and their signatures were obtained. The study was performed in accordance with the Declaration of Helsinki and approved by the ethics committee of Akdeniz University (approval # date: 12.03.2018, number: 70904504/111). The demographic, clinical characteristics, treatments, and colchicine doses of the patients and MEFV genotypes were evaluated. This is a cross-sectional study that was performed between January 2018 and February 2019. Laboratory results (complete blood count, Alanine aminotransferase (ALT), C - reactive protein (CRP), erythrocyte sedimentation rate (ESR), serum amyloid A (SAA), and analysing urine sample) were obtained from the medical records. Colchicine resistance was defined as the frequency of attacks remaining constant in patients receiving regular and adequate doses of colchicine therapy for at least 6 months or having at least one attack per month [11]. The colchicine usage dose of the patients was grouped in 2 different ways. One of the groupings was as patients using less than or more than 1,2 mg/m2/day, and the other was to compare the dose of colchicine calculated according to the patients' body weight according to the variant distribution.
Variant analysis:
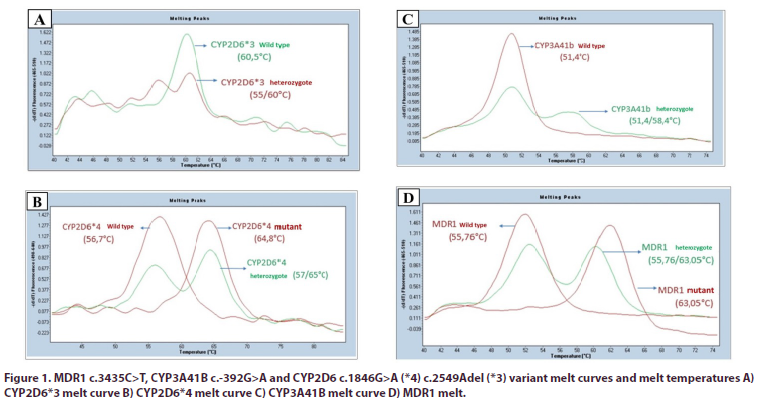
Genomic deoxyribonucleic acid (DNA) was obtained from the peripheral venous blood sample of the patients by silica column method, DNA quality and amount were determined by spectrophotometer, and DNA samples were stored at -20°C. As target gene regions, MDR1 c.3435C>T (rs1045642), CYP3A41B c.-392G>A (rs2740574), CYP2D6*4 c.1934G>A (rs3892097) and CYP2D6*3 c.2637Adel (rs35742686) variants were examined. The analysis of these variants was carried out using the melt curve method to be performed after amplification by quantitative PCR using fluorescent probes specific for alleles (Figure 1). Existing devices (LC480 Quantitative PCR Device) were used in the SBAUM infrastructure for quantitative PCR operations. The findings detected in the target genes evaluated were classified as wild type (WT), heterozygous (HET) and mutant (MUT) according to the variant distribution.
Statistical analysis:
Numerical data was expressed as mean, standard deviation, median, and categorical variables were expressed as percentages. The difference between categorical variables was evaluated with the chi-square test. The difference in the mean values between the groups was evaluated with the “Student t test” for data showing a normal distribution, and with the “Mann- Whitney U” and “Kruskal Wallis” tests for data showing a normal distribution. Statistical analyses of the data were performed using the SPSS 20.0 package program, and a value of p<0.05 was considered statistically significant. The distribution of alleles was evaluated by the chisquare test for compliance with the Hardy-Weinberg distribution. The statistical evaluation was performed by the Akdeniz University Statistical Advisory Unit.
Results
Of the 124 children included in the study, 55 were girls (44.4%) and 69 were boys. The mean age of the patients was 11.4±4.2 years, and the age at diagnosis was 6.3±3.7 years. The median follow-up period was 52.9 months. The mean age of the control group was 9.1±4.8 years (3-18 years), and the gender distribution was similar to the patient group. Thirty-five (28.2%) of the patients had a history of consanguineous marriage. Almost half of the patients (49.2%) had a first or second degree relative with a diagnosis of FMF. All patients were under colchicine treatment (preparations available in Turkey; Colchicum-dispert®, Kolsin®). 34 of the patients (27.4%) were receiving colchicine >1.2 mg/m2/day and their disease was under control. Colchicine resistance was present in 16 patients (12.9%). 5 of these patients were using the magnesium-containing form of colchicine (Colchicina lirca® and Colchicina opacalcium®), 5 of them were under canakinumab treatment. The results are summarized on (Table 1).
| Demographic and clinical characteristics of the patient group | |
|---|---|
| Age (mean ± SD) | 11,41± 4,2 years |
| Female gender (n/%) | 55 (44,4%) |
| Age at onset of treatment (mean ± SD) | 6,3±3.69 (1-15) years |
| Diagnostic delay (mean ± SD) | 11.3±23.556 months |
| Follow-up duration (mean ± SD) | 56.5±52.9 months |
| Last Control Age (mean ± SD) | 10.67±4.543 (3-18) years |
| Consanguineous marriage (n/%) | 35 (28,2%) |
| Family history of FMF (n/%) | 61 (49,2%) |
| Colchicine resistance (n/%) | 16 (12,9%) |
| Treatments received by resistant patients | 5 |
| Canakinumab (n) | |
| Other colchicine preparations (n) | 5 |
| Colchicine Dose (n/%) | 34 (27,4%) |
| >1,2 mg/m2/day | |
| <1,2 mg/m2/day | 90 (72,6%) |
| mg/kg/day (mean ± SD) | 0,0346 ± 0,015 |
| Frequency of side effects (n/%) | 15 (12,1%) |
| Diarrhea | 9 (7,3%) |
| Nausea – vomiting | 2 (1,6%) |
| Leukopenia | 2 (1,6%) |
| Thrombocytopenia | 2 (1,6%) |
| ALT elevation | 1 (0,8%) |
| Anemia | 1 (0,8%) |
| Hair loss | 1 (0,8%) |
Abdominal pain was the most common complaint among the reasons for admission, and fever was the second most common complaint. Joint complaints (arthritis, arthralgia), erythema, myalgia, pleurisy were other complaints at admission. The results are summarized on (Table 2). Of the patients, 50 (40.3%) were homozygous, 47 (37.9%) were compound heterozygous, and 27 (21.8%) had a heterozygous MEFV gene mutation. The most common MEFV mutation was M694V homozygous, and the most common MEFV gene mutation in the colchicine resistant group was M694V homozygous (62.5%). Genetic data was summarized in (Table 3).
| Complaints and clinical findings | Number of patients n (%) |
|---|---|
| Fever | 102 (82,3%) |
| Abdominal pain | 104 (83,9%) |
| Joint involvement | 80 (64,5%) |
| Erythema | 14 (11,3%) |
| Myalgia | 12 (9,7%) |
| Pleurisy | 6 (4,8%) |
| Livedo reticularis | 2 (1,6%) |
| Nausea-vomiting | 2 (1,6%) |
| Appendectomy history | 2 (1,6%) |
| Amyloidosis | 3 (2,4%) |
| Splenomegaly | 6 (4,8%) |
| Proteinuria | 12 (9,7%) |
| Growth retardation (Height and/or weight <3p) | 14 (11,3%) |
| Mutation distribution | Mutation | Number |
|---|---|---|
| Homozygous (n=50, 40.3%) | M694V/M694V R202Q/R202Q M680I /M680I E148Q/E148Q V726A /V726A E167D/E167D | 40 5 2 1 1 1 |
| Compound heterozygous (n=47, 37.9%) | M694V/R202Q M694V/M680I M694V/M680I/R202Q M694V/R202Q/V726A M694V/R761H M694V/V726A E148Q/P369S P369S/R202Q M694V/E148Q E148Q/R202Q A744S/R202Q A744S/P369S M680I/R202Q E148Q/R202Q/P369S M694V/P369S/R202Q P369S/A744S/R202Q E176D/M694V/R202Q | 23 3 2 2 2 2 2 1 2 1 1 1 1 1 1 1 1 |
| Heterozygous (n=27, 21.8%) | M694V/N E148Q/N M680I/N V726A/N R202Q/N | 16 6 3 2 1 |
MDR1 variants and alleles were similar in study and control groups (p=0.838). No significant relationship was found between MDR1 variant, allele distribution and colchicine resistance. While 80% of the patients in the TT (mutant) variant were in the low-dose colchicine (<1.2 mg/m2/day) group, 68.2% of the patients in the CC (mutant) variant were receiving low-dose colchicine and there was a statistically significant difference between those with T allele and C allele (p=0.046). When the colchicine doses were assessed in mg/kg/day, the mean dose was found to be 0.036mg/kg/day in the CC (wild type) variant, 0.033mg/kg/day in the CT (heterozygous) and TT variants, which was not statistically significant (p=0.56). The results were summarized on (Table 4).
| Colchicine dose mg/kg/m2 | p | Colchicine dose mg/kg/day | p | Colchicine resistance | p | |||
|---|---|---|---|---|---|---|---|---|
| >1,2 | <1,2 | Mean ± SD | Yes (n=16) | No (n=108) | ||||
| CC variant (n, %) | 15 (34.1) | 29 (65.9) | 0.110 | 0.036 ± 0.03 | 0.560 | 5 (11.4) | 39 (88.6) | 0.911 |
| CT variant (n, %) | 16 (28.1) | 41 (71.9) | 0.033 ± 0.02 | 8 (14.0) | 49 (86) | |||
| TT variant (n, %) | 2 (9.5) | 19 (90.5) | 0.033 ± 0.04 | 3 (14.3) | 18 (85) | |||
| C allele (n, %) | 46 (31.7) | 99 (68.2) | 0.046 | 18 (12.5) | 127 (87.5) | 0.698 | ||
| T allele (n, %) | 20 (20) | 79 (80) | 14 (14.2) | 85 (85.8) | ||||
Heterozygous (GA) changes of CYP3A41b variants were detected in 2.4% and 3.3% of the patient and control groups, respectively, and no mutant (AA) changes were found in either group. Colchicine resistance was present in 33% and 13% of patients with G and A alleles, respectively. Most of the patients with G allele (66%) were using colchicine at a dose of <1.2 mg/m2/day. The distribution between the groups was not statistically significant (p=0.619). Similarly, no significant correlation was found between the dose of colchicine according to body weight and the distribution of variants and alleles.
CYP2D6*4 in 92 patients and 37 healthy individuals was found to be 33.6%/3.2% and 21.6%/2.7%, respectively. The distribution was similar in both groups. No relationship was found between variant distribution of A and G alleles and colchicine resistance. Considering the daily dose of colchicine according to body weight, the mutant (AA) group was using 0.017mg/kg/day, the heterozygous (GA) group was using 0.034mg/kg/day, and the wild-type group (GG) was using colchicine 0.035mg/kg/day. It was determined that a lower dose of colchicine was used in the mutant group than in heterozygous and wild-type groups. It was found that colchicine dose (mg/kg/day), which was divided into percentiles, was lower in the Cyp2D6*4 mutant group compared to the wild type and heterozygous groups (p=0,011/ p=0,033) (Table 5). On the other hand, CYP2D6*3 was found as wild-type in all 129 individuals whose variant could be examined, and other alleles were absent.
| Colchicine dose mg/kg/day | CYP2D6*4 | Percentile | Median | Quarterly range (IQR) | 95% Confidence interval (CI) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 5 | 10 | 25 | 50 | 75 | 90 | 95 | |||||
| Wildtype (GG) | ,0179 | ,020 | ,0245 | ,0325 | ,0450 | ,0500 | ,0570 | 0.033 | 0.021 | 0.031 – 0.038 | |
| Heterozygous (AG) | ,0127 | ,0163 | ,0207 | ,0300 | ,0457 | ,0600 | ,0758 | 0.030 | 0.025 | 0.028 – 0.041 | |
| Mutant (AA) | ,0100 | ,0100 | ,0100 | ,0200 | . | . | . | 0,020* | - | 0.002 – 0.031 | |
Discussion
Considering several challenges of biological treatment options, we tried to predict colchicine resistance in children with FMF by analysing the colchicine metabolism related variants. Although we could not find a significant relationship between variant distribution and colchicine resistance, we showed that the dose of colchicine used was significantly lower in those with the MDR1 mutant allele and those with CYP2D6*4 mutation compared to other patients. When the MDR1, CYP3A41B, CYP2D6*4 and CYP2D6*3 variant distributions were considered, the distribution was found to be similar in the patient and control groups, in almost all the 124 pediatric patients with a diagnosis of FMF using colchicine and 60 healthy children who were similar in age and gender.
Colchicine is a drug with a very narrow therapeutic dose range, and it is known to show individual differences in terms of bioavailability. In addition, its metabolism changes because of drug-drug interaction. All these properties are thought to be due to genetic variation of the CYP450 family of enzymes and P glycoprotein (MDR1) molecules that are involved in the metabolism of colchicine between individuals, and studies show that colchicine can be involved in the development of resistance and effective dose regulation by affecting the blood level [12,13,14].
Various dosage ranges have been suggested for colchicine use in children, based on both surface area and body weight [15]. There is no consensus on this matter. In our study, we evaluated the colchicine usage dose in 2 ways; the first was to use less than or more than 1.2 mg/ m2/day, and the other was to describe it as mg/kg/day. The average daily dose of colchicine use in our patients was 0.0346 ± 0.015 mg/kg/day, and 90 (72.6%) of them used colchicine at a dose of <1.2 mg/m2/day. While there was no relationship between CYP3A41b and CYP2D6*3 variant distribution and colchicine usage dose, data suggesting that the disease of those with MDR1 and CYP2D6*4 mutant variants was controlled with lower doses of colchicine were obtained.
We found that 80% of patients with TT variant used colchicine <1.2 mg/m2/day in the dose relationship with MDR1 variant and allele distribution. Low-dose colchicine requirement was significantly more common in the T allele group than in the C allele group. The number of studies evaluating the relationship between colchicine dose and variation is limited. In a study by Dogruer et al., there was no significant relationship between the colchicine dose used and the CYP3A4 and MDR1 variants [8]. Another study examined the relationship between MDR1 variant distribution and blood digoxin levels, which play an important role in metabolism, and it was discovered that patients with the TT variant had lower P-glycoprotein levels than those with the CC variant, and their blood digoxin levels were higher [16].
The fact that the level of colchicine in the blood was not examined is one of the most important limitations of our study. However, similar to the current findings, considering that the level of colchicine in the blood is higher in the TT (mutant) variant, it is possible to conclude that lower doses of colchicine are sufficient to control the disease of these patients. CYP2D6*4 1934AA (mutant) is known as a slow metabolizer. Several studies evaluate the relationship between drug dose and CYP2D6*4 variant, especially in antidepressant drugs. In a study in which 1198 patients were evaluated when the relationship between antidepressant use and CY2PD6*4 was examined, it was seen that the dose of drug use was lower in those with slow metabolizers [17]. In our study, we discovered that the mutant allele's colchicine usage dose (mg/kg/day) was statistically significantly lower than other variants in the relationship between the colchicine usage dose and CYP2D6*4 variant distribution (p=0,011/ p=0,033). Because the mutant AA variant is a slow metabolizer, it may provide higher blood colchicine levels, suggesting that a lower colchicine dose in this group may be sufficient for effective treatment. This finding should be confirmed by studies in which the blood level of colchicine is examined.
Similar to the literature 12, 12.9% of the patients had colchicine resistance in our study. In a study carried out in Turkey, the relationship between colchicine resistance and CYP2D6*1, *3, *4, *5 and *6 was evaluated. The study included 60 patients with colchicine resistance and 30 patients with colchicine response, and it was discovered that the frequency of CYP2D6 *4 and *6 mutant alleles was higher in the colchicine unresponsive group [9]. Conflicting findings were presented in several studies evaluating the relationship between MDR1 3435C>T variant distribution and colchicine response. The association of colchicine resistance with the C allele was significant in a study involving 120 adult FMF patients from our country, and the colchicine response was better in the TT allele [18].
Dogruer et al. evaluated the relationship between CYP3A4 and MDR1 variants and dose of colchicine, and found no significant difference [8]. Another study examined the distribution of the MDR1 3435C>T variant and the relationship between lymphocyte colchicine level and colchicine response in 58 colchicineresistant and 47 colchicine-responsive patients. The level of colchicine in lymphocytes was lower in the resistant group, but no relationship was found between the blood level of colchicine and the distribution of the C and T alleles. In terms of allele frequency, in contrast to other studies, it has been shown that colchicine resistance is associated with the TT variant [19]. It was compatible with the study that showed that the presence of the TT allele and the expression of the MDR1 gene product, P-glycoprotein, were inversely proportional [16]. In our study, when the distribution of MDR1, CYP3A41B and CYP2D6*3 and*4 variants was examined in patients with colchicine resistance, it was seen that there was no difference between the groups.
In a study by Rustemoglu et al., they presented the distribution of MDR1 alleles in the Turkish population and the differences in allele distribution between FMF patients and control patients. They showed that the MDR1 heterozygous change was significantly higher in the FMF group and suggested that this genotype may play a role in pathogenesis [10]. However, in our study, when the distribution of MDR1, CYP3A41B, CYP2D6*4 and CYP2D6*3 variants was evaluated in 124 children with FMF using colchicine and 60 healthy children who were similar in age and gender, the distribution was found to be similar. One of the most important limitations of the study is that the level of colchicine in the blood has not been studied. Another limitation is that there are few colchicine-resistant groups due to the limited number of participants in the study group, which consists entirely of pediatric patients.
Conclusion
No correlation was determined between colchicine resistance and disease severity and MDR1, CYP3A4, CYP2D6*3 and *4 variant distributions. However, lower doses of colchicine were used in the MDR1 and CYP2D6*4 mutant groups. Changes in the MDR1, CYP3A4, CYP2D6*3, and *4 variants were detected similarly in the patient and control groups. These variant changes' clinical significance and effect are revealed only in dosing medications. It is not cost-effective to perform this variant analysis before treatment.
However, it can be utilized to guide patients when adjusting their individualized treatment dose.
Availability of data and material
All of the data are available on request from the authors.
Limitations
One of the most important limitations of the study is that the level of colchicine in the blood has not been studied. Another limitation is that there are few colchicineresistant groups due to the number of the study group, which consists entirely of pediatric patients.
Conflicts of Interest
All the authors declare no conflict of interest related to the study.
References
- Ozen S, Bilginer Y. A clinical guide to autoinflammatory diseases: Familial Mediterranean fever and next-of-kin. Nat Rev Rheumatol. 10(3), 135-47 (2014).
- Tufan A, Lachmann HJ. Familial Mediterranean fever, from pathogenesis to treatment: a contemporary review. Turk J Med Sci. 50(SI-2),1591-1610 (2020).
- Ozen S, Kone-Paut I, Gül A. Colchicine resistance and intolerance in familial mediterranean fever: Definition, causes, and alternative treatments. Semin Arthritis Rheum. 47(1), 115-20 (2017).
- Dasgeb B, Kornreich D, McGuinn K et al. Colchicine: an ancient drug with novel applications. Br J Dermatol. 178(2), 350-6 (2018).
- Phillips KA, Veenstra DL, Oren E et al. Potential role of pharmacogenomics in reducing adverse drug reactions: a systematic review. JAMA. 286(18), 2270-9 (2001).
- Lim HS, Lee HJ, Lee KS et al. Clinical implications of CYP2D6 genotypes predictive of tamoxifen pharmacokinetics in metastatic breast cancer. J Clin Oncol. 25, 3837-45 (2007).
- Tufan A, Babaoglu MO, Akdogan A et al. Association of drug transporter gene ABCB1 (MDR1) 3435C to T polymorphism with colchicine response in familial Mediterranean fever. J Rheumatol. 34(7), 1540-4 (2007).
- Dogruer D, Tug E, Bes C et al. Lack of an effect of CYP3A4 and MDR1 gene polymorphisms on colchicine pharmacogenetics in the treatment of Familial Mediterranean fever. Genet Mol Res. 12(3), 3521-8 (2013).
- Yalcıntepe S, Ozdemır O, Sılan C et al. The CYP4502D6 *4 and *6 alleles are the molecular genetic markers for drug response: implications in colchicine non-responder FMF patients. Eur J Drug Metab Pharmacokinet. 41(3), 281-6 (2016).
- Demirkaya E, Acikel C, Hashkes P et al. Development and initial validation of international severity scoring system for familial Mediterranean fever (ISSF). Ann Rheum Dis. 75(6), 1051-6 (2016).
- Ozen S, Demirkaya E, Erer B et al. EULAR recommendations for the management of familial Mediterranean fever. Ann Rheum Dis. 75(4), 644-51 (2016).
- Ben-Chetrit E, Aamar S. About colchicine compliance, resistance and virulence. Clin Exp Rheumatol. 27(2 Suppl 53), S1-3.
- Dahan A, Sabit H, Amidon GL. Multiple efflux pumps are involved in the transepithelial transport of colchicine: combined effect of p-glycoprotein and multidrug resistance-associated protein 2 leads to decreased intestinal absorption throughout the entire small intestine. Drug Metab Dispos. 37(10), 2028-36 (2009).
- Tateishi T, Soucek P, Caraco Y et al. Colchicine biotransformation by human liver microsomes. Identification of CYP3A4 as the major isoform responsible for colchicine demethylation. Biochem Pharmacol [Internet]. 53(1), 111-6 (1997).
- Ozkaya N, Yalcinkaya F. Colchicine treatment in children with familial Mediterranean fever. Clin Rheumatol. 22(4-5), 14-7 (2003).
- Hitzl M, Drescher S, van der Kuip H et al. The C3435T mutation in the human MDR1 gene is associated with altered efflux of the P-glycoprotein substrate rhodamine 123 from CD56+ natural killer cells. Pharmacogenetics. 11(4), 293-8 (2001).
- Bijl MJ, Visser LE, Hofman A et al. Influence of the CYP2D6*4 polymorphism on dose, switching and discontinuation of antidepressants. Br J Clin Pharmacol. 65(4), 558-64 (2008).
- Bezalel Y, Gershoni-Baruch R, Dagan E et al. The 3435T polymorphism in the ABCB1 gene and colchicine unresponsiveness in familial Mediterranean fever. Clin Exp Rheumatol. 27(2 Suppl 53), S103-4 (2009).
- Rüstemoglu A, Gümüş-Akay G, Yigit S et al. Analysis of common MDR1 (ABCB1) gene C1236T and C3435T polymorphisms in Turkish patients with familial Mediterranean fever. Genet Mol Res. 10(4), 3411-20 (2011).
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref