Special Issue Article - Imaging in Medicine (2011) Volume 3, Issue 4
Tracking stem cells for cardiovascular applications in vivo: focus on imaging techniques
Yingli Fu1, Nicole Azene1,2, Yi Xu1 and Dara L Kraitchman†1,2
1 Russell H Morgan Department of Radiology & Radiological Science, Johns Hopkins University, Baltimore, MD, USA
2Department of Molecular& Comparative Pathobiology, Johns Hopkins University, Baltimore, MD, USA
- *Corresponding Author:
- Dara L Kraitchman
Russell H Morgan
Department of Radiology & Radiological Science
Johns Hopkins University, Baltimore, MD, USA
Tel.: +1 410 955 4892
Fax: +1 410 614 1977
E-mail: dkraitc1@jhmi.edu
Abstract
Despite rapid translation of stem cell therapy into clinical practice, the treatment of cardiovascular disease using embryonic stem cells, adult stem and progenitor cells or induced pluripotent stem cells has not yielded satisfactory results to date. Noninvasive stem cell imaging techniques could provide greater insight into not only the therapeutic benefit, but also the fundamental mechanisms underlying stem cell fate, migration, survival and engraftment in vivo. This information could also assist in the appropriate choice of stem cell type(s), delivery routes and dosing regimes in clinical cardiovascular stem cell trials. Multiple imaging modalities, such as MRI, PET, SPECT and CT, have emerged, offering the ability to localize, monitor and track stem cells in vivo. This article discusses stem cell labeling approaches and highlights the latest cardiac stem cell imaging techniques that may help clinicians, research scientists or other healthcare professionals select the best cellular therapeutics for cardiovascular disease management.
Keywords
cardiovascular disease; cell labeling; cell tracking; computed tomography; MRI; optical imaging; radionuclide imaging; stem cells
Coronary heart disease is usually a result of atherosclerotic progression, which is characterized by the narrowing of small blood vessels that supply blood and oxygen to the heart. Despite recent advances in pharmacotherapy and interventional procedures, coronary heart disease remains the leading cause of heart failure in the western world [1]. Owing to the limited regenerative capacity of cardiac cells and subsequent detrimental ventricular remodeling after myocardial infarction, heart transplantation is currently the only definitive treatment for end-stage cardiovascular disease. Yet, the lack of suitable organs and high cost limit the number of heart transplants that are performed. Stem and progenitor cells possess the capability of self-renewal and can differentiate into organ-specific cell types with the potential to reconstitute damaged organ systems. Thus, many types of stem cell therapeutics have emerged for cardiac repair with rapid translation to clinical trials, sometimes without extensive preclinical testing.
Preclinical cardiovascular stem cell studies in animal models of myocardial infarction have shown encouraging results ranging from restoration of ventricular function to improved myocardial perfusion [2]. The results from clinical trials, however, have been mixed (Table 1). The BOOST trial demonstrated the safety and feasibility of intracoronary infusion of autologous bone marrow cells and early improvements of left ventricular function [3], but the improvements were not sustained after 5 years [4]. In addition, other double-blinded, randomized and placebo-controlled trials showed little or no long-term benefits [5–7]. Each clinical trial has involved a relatively small number of patients with a primary end point of safety. However, meta-analyses of these trials have generally shown a slight positive trend towards improved cardiac function [8–12].

Noninvasive imaging, such as echocardiography, MRI, PET, SPECT and CT, can play a pivotal role in tracking stem cell engraftment and expand these imaging modalities beyond merely assessing cardiac function. This article will provide an overview of the fundamentals of stem cell labeling techniques and discuss the advantages and disadvantages of each imaging modality with a focus on those with the greatest potential for clinical translation.
Stem cell labeling
Embryonic stem cells (ESCs), adult stem cells and induced pluripotent stem cells are the three major types of stem/progenitor cells that possess the ability to provide the building blocks for cardiovascular system repair. Owing to the ethical issues and the potential danger of teratoma formation of undifferentiated ESCs and the low reprogramming efficiency of induced pluripotent stem cells, adult stem cells (e.g., bone marrow-derived mesenchymal stem cells, skeletal myoblasts and cardiac/endothelial progenitor cells) have dominated current clinical investigations (Table 1).
For noninvasive tracking of stem cells, the label should ideally be [13]:
▪ Inert and/or biocompatible
▪ Highly specific to target cells
▪ Detected at the level of a single or few cells
At present, there is no single label/probe that meets all these requirements in combination with a single imaging technique. However, each imaging modality has desirable characteristics that, in part, may drive the choice of stem cell labels (Table 2). In general, stem cell labeling falls into two primary methods: direct/physical labeling and indirect/genetic labeling.

Direct cell labeling typically requires incubation of a cell with the label of interest. The label may then be bound on the cell surface or internalized by the cell. As such, direct labeling is the simplest and most straightforward method that can be performed with a variety of probes to enable visualization of stem cells by noninvasive clinical imaging techniques [14]. For example, direct stem cell labeling using superparamagnetic iron oxide particles (SPIOs) and radioactive tracers (e.g., 111In and 18F) has been widely used for MRI [15–17] and radionuclide imaging [18,19], respectively. Optical imaging of stem cells labeled with fluorescence probes (e.g., near-infrared fluorophores and quantum dots) has been reported in animal models [20,21]. The primary disadvantages of direct labeling are label dilution/loss with cell division or cell death and inability to differentiate viable from dead cells due to retained label in situ or native cell uptake of label from dead cells. Thus, direct cell labeling is best for confirmation of cell delivery success and short-term localization of cells after delivery.
In contrast to direct stem cell labeling, genetic labeling of stem cells with reporter genes involves insertion of genetic material that encodes for an enzyme, receptor or protein that can then be imaged directly or interacts with a reporter probe to enable noninvasive visualization of the cell. Because only live cells can produce the reporter gene product, this technique is better at discriminating live from dead cells and, thus, can provide longitudinal imaging of cell survival and engraftment in vivo. Several reporter genes, such as firefly luciferase (Fluc, bioluminescence imaging reporter) [22], herpes simplex virus thymidine kinase (HSVtk, PET reporter) [22] and ferritin (MRI reporter) [23] have been constructed for stem cell tracking on different imaging platforms. However, safety concerns regarding genetic cell manipulation have limited reporter gene labeling clinical adoption.
Echocardiography
Owing to its wide availability, low cost and lack of ionizing radiation, echocardiography is routinely used to assess cardiac function, diagnose pericardial disease and evaluate stem cell therapy efficacy. However, few studies have explored using echocardiography to track cells. In part, this is due to the low spatial resolution and lack of accuracy in cell quantification of ultrasound. Cardiac stem cell tracking using microbubbles [24,25] or CliniMACS® nanoparticles [26] have very recently been performed. In one study, the engraftment of genetically modified endothelial progenitor cells within a Matrigel™ plug was imaged with contrast-enhanced ultrasound using targeted microbubbles [25]. Two issues with microbubble technology are that the microbubble integrity cannot be maintained over time and the delivery of microbubbles outside the vascular space is challenging.
MRI
MRI is a multipurpose imaging modality that provides excellent soft tissue contrast with high spatial and moderate temporal resolution. Therefore, it has been frequently used in the clinic to assess cardiac anatomy, ventricular function, blood flow and myocardial perfusion. In noncardiac applications, MRI has been used to visualize individual labeled cells against a homogeneous background [27]. Stem cells can be directly labeled with a magnetic resonance (MR) contrast agent (e.g., gadolinium chelates and SPIOs) through endocytosis, magnetofection [28] or electroporation [29,30] approaches, or indirectly labeled with an MR reporter gene (e.g., ferritin) via viral [31] or nonviral transfection [32]. Thus, MRI offers the ability not only to determine the efficacy of stem cells, but to track the engraftment of stem cells based on the local environment using a single imaging modality.
Gadolinium chelates
As the first US FDA-approved MR contrast agents, gadolinium-chelated contrast agents have been widely used in off-label approaches to quantify myocardial perfusion and viability [33]. On T1-weighted MRI, cells labeled with paramagnetic gadolinium chelates appear hyperintense. Recently, a gadolinium-based contrast agent, Cy3-labeled gadofluorine M, has been used to label ESC-derived cardiac progenitor cells [34]. No effect on cell viability was observed in vitro and transplanted cells could be imaged in vivo 2 weeks post injection in both infarcted and normal mice [34]. Interestingly, this agent overcame the issue of intracellular compartmentalization of gadolinium in labeled cells, which results in smaller decreases in T1 relaxivity or decreased sensitivity to gadolinium-labeled cells [35]. Nonetheless, since unchelated gadolinium is highly toxic, concerns about clinical utilization of these agents for cell tracking remain.
Superparamagnetic iron oxides
Superparamagnetic iron oxide nanoparticles are the most widely used MR contrast agents for cellular labeling and typically impart hypointense contrast on T2*-weighted images [36]. For the most part, direct labeling of stem cells with SPIOs is performed prior to cell transplantation. Because most stem cells of interest for cardiovascular applications are not phagocytic cells, spontaneous uptake of SPIOs is unlikely to occur. Instead, a positively charged transfection agent, such as poly-l-lysine, cationic liposome or protamine sulfate, is often used to coat the negatively charged iron oxide particles to enable cellular uptake of the transfection agent–SPIO complex through electrostatic interactions. Once within the cells, extremely small quantities of SPIOs (e.g., picograms) can generate high contrast without the toxicity concerns of gadolinium since SPIOs are biodegradable and recycled into the normal iron pool [37].
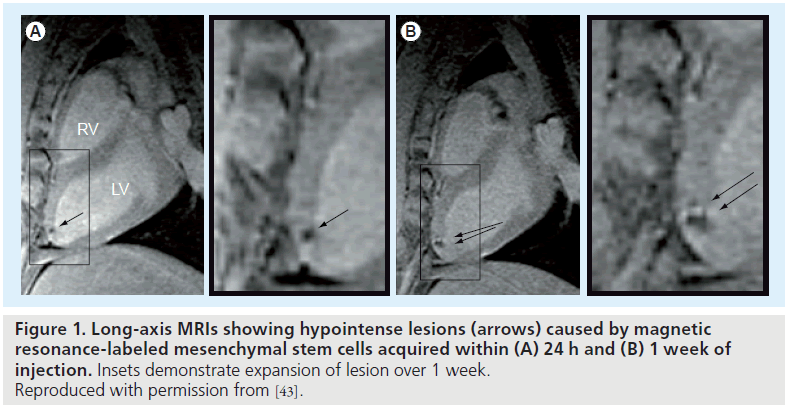
To date, no clinical trials using SPIO-labeled stem cells have been initiated for cardiac repair. However, several studies with tracking of SPIO-labeled stem cells have now been performed in other diseases [30,38–41]. In preclinical cardiovascular applications, MR-based tracking of SPIO-labeled ESCs, mesenchymal stem cells (MSCs) and hematopoietic stem cells (HSCs) have been performed in varied animal models [42–44]. In one study of SPIO-labeled mouse ESCs, the hypointensities in the ischemic myocardium were observed 4 weeks after implantation in mice, indicating the successful integration of labeled ESCs with infarcted myocardium [42]. In addition, intramyocardially injected SPIOlabeled MSCs could be detected by MRI up to 3 weeks after infarction in pigs (Figure 1) [43]. A similar study carried out by Amado et al. demonstrated substantial retention of SPIO-labeled bone marrow-derived stromal cells in infarcted myocardium at 8 weeks [45].
Figure 1. Long-axis MRIs showing hypointense lesions (arrows) caused by magnetic resonance-labeled mesenchymal stem cells acquired within (A) 24 h and (B) 1 week of injection. Insets demonstrate expansion of lesion over 1 week. Reproduced with permission from [43].
In an effort to take additional advantage of the lack of ionizing radiation with MRI and the ability to see the success of injections immediately, several groups have developed MR-compatible delivery devices often in combination with graphical interfaces for real-time MRI to perform MR-based stem cell delivery interventions [46–48]. At present, the regulatory hurdles for labeling stem cells are sufficiently large such that the addition of an investigational new device for delivery on an MRI platform that is not familiar to interventional cardiologists will probably limit the adoption of these techniques clinically in the near future.
Furthermore, beyond the normal limitations of direct cell labeling, such as possibility of signal dilution with cell replication and detachment of the label from the cell [49,50], the signal void generated by SPIOs may be problematic to distinguish from native tissue hypointensities, such as areas of ischemia, calcification and hemorrhage, motion artifacts and the presence of metallic objects (e.g., stents). To tackle this problem, a variety of positive- contrast MRI techniques or post-imaging processing methods (e.g., IRON, SWIFT and PARTS) [44,51–53] have been developed. However, the biggest hurdle to clinical adoption of these techniques is the removal of commercially available SPIO formulations. Nevertheless, preclinical studies of cardiac SPIO stem cell tracking will continue to be an active field that can be used to help guide clinical trials with respect to dosing, timing and cell choices with the caveat that any direct labeling scheme, such as SPIOs, may become detached from the cell of interest.
Other MR contrast agents
In addition to 1H-based contrast labeling agents, other nonproton-based compounds containing 19F, 23Na or 13C have also been explored for MR detection. In particular, 19F-based agents have been used by several investigators for stem cell tracking [54–57]. Because there is essentially no native fluorine in the body, 19F ‘hot spot’ imaging [58] can be achieved with high sensitivity. However, specialized hardware and MRI sequences are often required to perform such studies.
MR reporter genes
MR reporter gene labeling may address the problems associated with direct MR contrast labeling. Several MR reporters have been developed, including creatine kinase [59], iron storage proteins (e.g., ferritin, transferrin and transferrin receptor) [23,32,60–63] and artificial proteins (e.g., lysine-rich protein) [64]. Overexpression of the transgenic human ferritin receptor and ferritin heavy chain subunit has been induced in tumor cells [32], neural stem cells [62] and ESCs [60]. Cardiac applications, however, have not yet been explored. Recently, preclinical studies have demonstrated the feasibility of labeling mouse skeletal myoblasts with the MR reporter ferritin. These transgenic cells were successfully detected by MRI in vitro and in vivo after transplantation into the infarcted mouse heart [23]. Besides safety concerns due to genetic alteration, the primary inherent problem with imaging of MR reporters is whether small numbers of cells can generate sufficient reporter gene products to enable visualization.
Radionuclide imaging
Radionuclide imaging, including PET and SPECT, has the highest sensitivity (PET: 10-11 to 10-12 mol/l; SPECT: 10-10 to 10-11 mol/l) and spatial resolution among all currently used imaging modalities with the ability to quantify radioisotope levels [65]. Clinically, radionuclide imaging has been routinely used to assess cardiac metabolic function, viability, contractile function, as well as cell tracking [66,67]. In general, radioisotopes with a relatively long decay half-life are preferred for cell labeling and tracking.
Direct radiolabeling
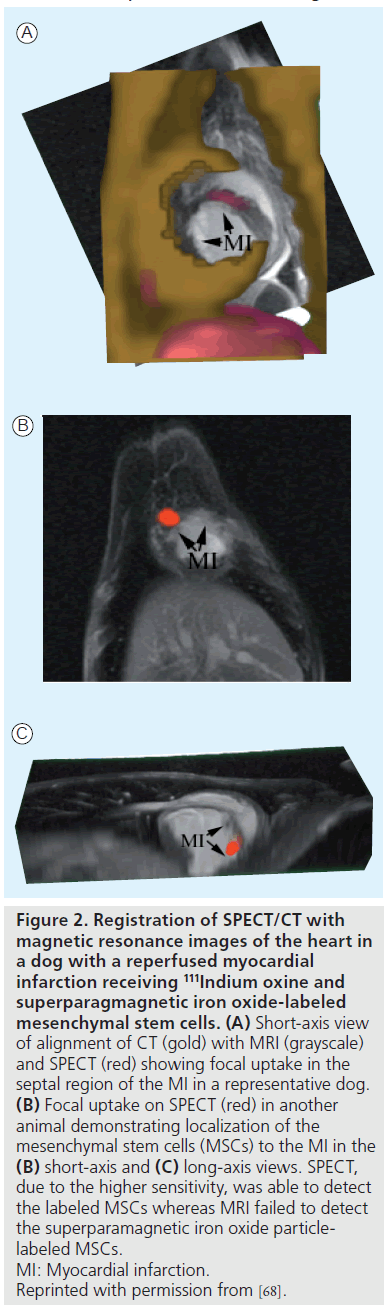
Direct cell labeling with 111In oxine (t1/2 ≈ 2.8 days) developed for lymphocyte labeling has been adapted for cardiac stem cell imaging. For example, the trafficking of 111In oxine and SPIO-labeled MSCs could be monitored by clinical SPECT/CT up to 7 days in the infarcted canine myocardium, while MRI failed to detect the cells (Figure 2) [68]. Several other studies have demonstrated the varied retention of radiolabeled stem cells in the heart depending on the route of delivery, such as intravenous, intramyocardial, intracoronary and interstitial retrograde coronary venous delivery [69–76]. 111In has also been used in patient studies to better understand the trafficking of peripheral blood progenitor cells after intracoronary and intravenous delivery in patients with heart disease [77,78]. 18F-FDG (t1/2 ~110 min) is another attractive radiotracer that is more readily available for stem cell labeling. The first human study demonstrated higher retention of 18F-FDGlabeled CD34-positive enriched bone marrow mononuclear cells in the infarcted myocardium than nonselected bone marrow cells 70 min after intracoronary delivery [79]. In a similar study, PET imaging revealed less than 3.3% of 18F-FDGlabeled HSCs accumulated within the infarcted myocardium at 2 h post-delivery [80]. Similar to MRI, which can provide viability and anatomical location information as well as cell tracking, dual isotope imaging, such as 18F-FDG or 99mTc with 111In, may be used to monitor cell migration relative to local tissue perfusion or metabolism [70,81].
Figure 2. Registration of SPECT/CT with magnetic resonance images of the heart in a dog with a reperfused myocardial infarction receiving 111Indium oxine and superparagmagnetic iron oxide-labeled mesenchymal stem cells. (A) Short-axis view of alignment of CT (gold) with MRI (grayscale) and SPECT (red) showing focal uptake in the septal region of the MI in a representative dog. (B) Focal uptake on SPECT (red) in another animal demonstrating localization of the mesenchymal stem cells (MSCs) to the MI in the (B) short-axis and (C) long-axis views. SPECT, due to the higher sensitivity, was able to detect the labeled MSCs whereas MRI failed to detect the superparamagnetic iron oxide particlelabeled MSCs. MI: Myocardial infarction. Reprinted with permission from [68].
Depending on the radiotracer used and cell type, minimum detection limits of direct radiotracer labeling vary from 2900 to 25,000 cells [82]. Major concerns of direct radiotracer labeling include the potential radiation damage to the cells [82–84], leakage of radiotracers over the time course [85], short imaging window due to radioactivity decay and radiotracer detachment from the cells such as direct MRI contrast labeling.
PET/SPECT reporter gene labeling
To date, three major PET/SPECT reporter genes, namely enzyme based, receptor based and transporter based, have been developed and applied to cardiac imaging in large animals or human studies. Examples include transporterbased sodium-iodide symporter (NIS) [86–88] for SPECT imaging, receptor-based dopamine type 2 receptor (D2R) [89,90] and the most commonly used enzyme-based herpes simplex virus type 1 thymidine kinase (HSV1-tk) or its mutant form HSV1-sr39tk [22] for SPECT/PET imaging.
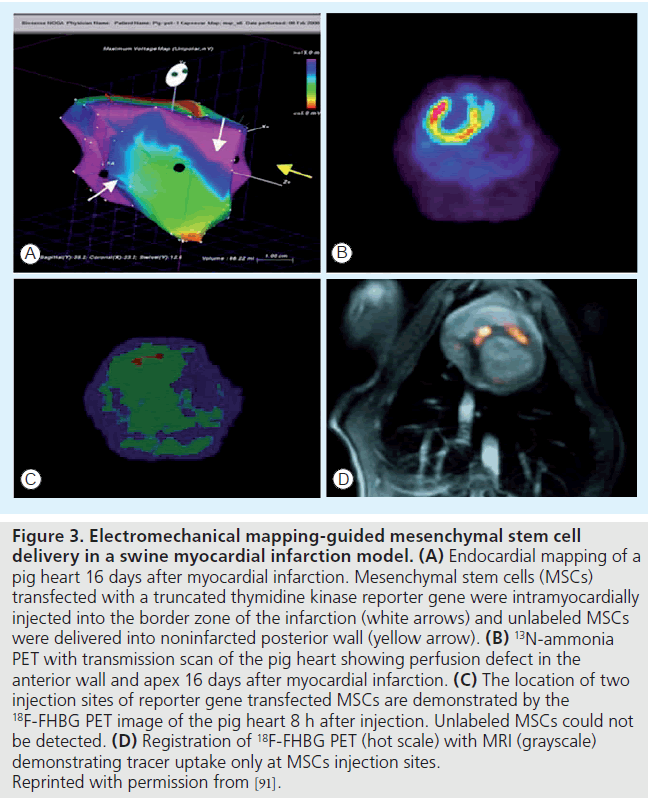
The reporter probes for imaging thymidine kinase reporter genes are radiolabeled pyrimidine nucleoside analogs (such as 18F-FHBG and 123I-FIAU/124I-FIAU) and acycloguanosine. In a large animal model of myocardial infarction, Gyöngyösi et al. demonstrated the first successful translation of PET imaging of the HSV1-tk reporter gene to track cardiac stem cell biodistribution after intramyocardial injection using electromechanical mapping guidance (Figure 3) [91]. Enzyme-based PET reporter gene labeling has the advantage of signal amplification. Thus, a very low level of reporter gene expression or small number of transplanted cells can often be detected using radionuclide imaging. The major limitations include potential immune response elicitation to the foreign reporter gene product, limited reporter probe trapping due to rate limited probe transport into the cells and silencing of the reporter gene leading to inability to detect the transplanted cells [92]. In addition, leakage of the reporter probe from cells transfected with the reporter gene has also been reported with the NIS reporter gene [88]. Although radionuclide imaging shows a high sensitivity to a small number of cells, anatomical information is lacking. Thus, CT or MRI is needed to provide localization of cell distribution.
Figure 3. Electromechanical mapping-guided mesenchymal stem cell delivery in a swine myocardial infarction model. (A) Endocardial mapping of a pig heart 16 days after myocardial infarction. Mesenchymal stem cells (MSCs) transfected with a truncated thymidine kinase reporter gene were intramyocardially injected into the border zone of the infarction (white arrows) and unlabeled MSCs were delivered into noninfarcted posterior wall (yellow arrow). (B) 13N-ammonia PET with transmission scan of the pig heart showing perfusion defect in the anterior wall and apex 16 days after myocardial infarction. (C) The location of two injection sites of reporter gene transfected MSCs are demonstrated by the 18F-FHBG PET image of the pig heart 8 h after injection. Unlabeled MSCs could not be detected. (D) Registration of 18F-FHBG PET (hot scale) with MRI (grayscale) demonstrating tracer uptake only at MSCs injection sites. Reprinted with permission from [91].
Optical imaging
Optical imaging techniques, including fluorescence imaging and bioluminescence imaging, can provide high sensitivity for cell tracking with detectability of 10-9 to 10-12 mol/l and 10-15 to 10-17 mol/l, respectively [93]. In particular, optical imaging of reporter genes (e.g., green fluorescence protein or luciferase) can be used to monitor stem cell survival, proliferation and cardiac-specific differentiation in small animals [49,94]. However, technical challenges, such as the limited tissue penetration and low energy photon attenuation that restricts visualization of deep structures, such as blood vessels and the heart, limit development of clinical imaging systems [95,96].
Multimodality imaging
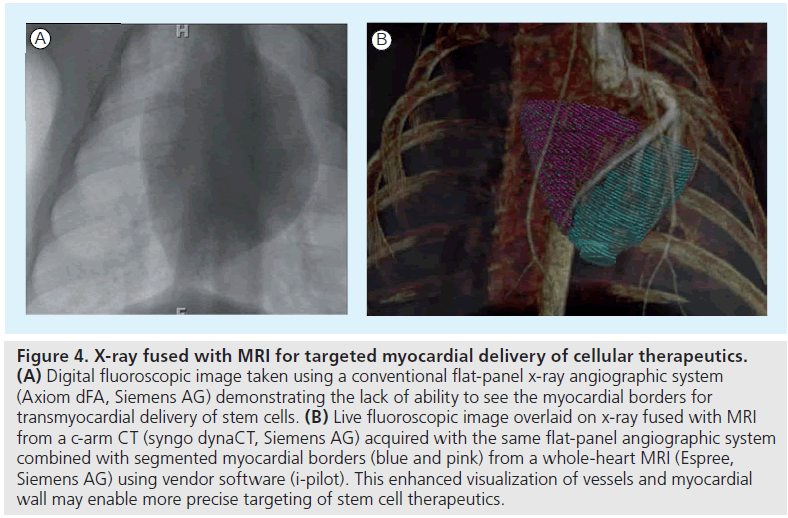
Multimodality imaging, such as the combination of CT with SPECT or PET, to obtain high sensitivity and anatomical detail can be expanded to other technologies to enhance stem cell tracking and measurement of cardiovascular function. A solution to some of the disadvantages of MRI, such as lack of MR compatible devices, poor physiological monitoring and limited temporal resolution for real-time interventions, would be to combine MRI with x-ray interventional techniques for stem cell delivery (Figure 4). Fusion of myocardial anatomy and viability maps from MRI in a swine infarction model have been used to target injections to the infarct borders using an x-ray fused with a MR registration platform [97].
Figure 4. X-ray fused with MRI for targeted myocardial delivery of cellular therapeutics. (A) Digital fluoroscopic image taken using a conventional flat-panel x-ray angiographic system (Axiom dFA, Siemens AG) demonstrating the lack of ability to see the myocardial borders for transmyocardial delivery of stem cells. (B) Live fluoroscopic image overlaid on x-ray fused with MRI from a c-arm CT (syngo dynaCT, Siemens AG) acquired with the same flat-panel angiographic system combined with segmented myocardial borders (blue and pink) from a whole-heart MRI (Espree, Siemens AG) using vendor software (i-pilot). This enhanced visualization of vessels and myocardial wall may enable more precise targeting of stem cell therapeutics.
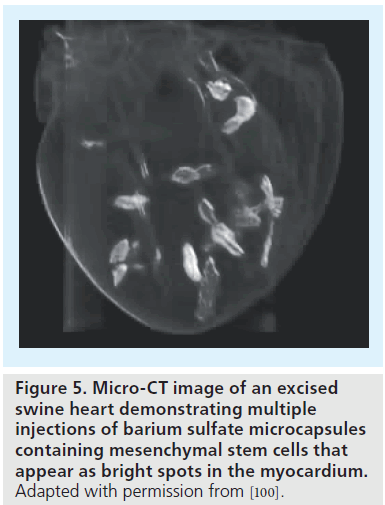
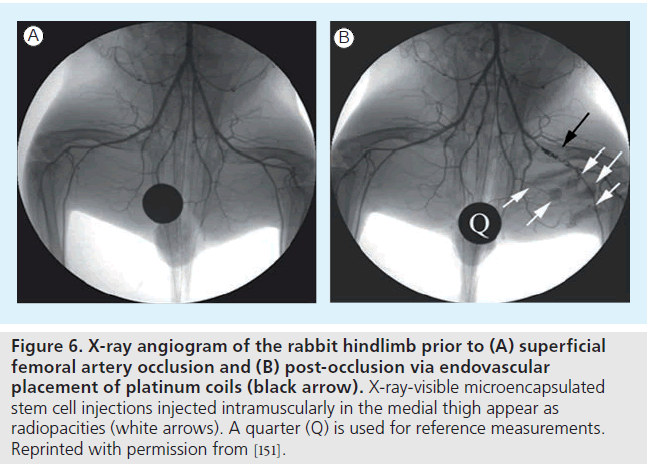
Although stem cell labeling enables noninvasive visualization of cells in infarcted or normal subjects, a big concern for cardiac stem cell therapy is the significant cell death, which may be attributed, in part, to the ischemic environment and immunodestruction. To overcome early cell death, a hybrid technique whereby cells are encapsulated in a protective barrier (which blocks cell destruction by immunoglobulins and immune-mediated cells) and impregnated with imaging contrast agents is currently being explored [55,98,99]. By moving the labeling agents to the protective capsule rather than within cells, high contrast payloads for enhanced sensitivity can be used, which would often be cytotoxic. Recently, these imaging-visible microcapsules have been demonstrated in a swine heart (Figure 5) [100] and also used to track stem cells in a rabbit model of peripheral arterial disease (Figure 6) [101].
Figure 5. Micro-CT image of an excised swine heart demonstrating multiple injections of barium sulfate microcapsules containing mesenchymal stem cells that appear as bright spots in the myocardium. Adapted with permission from [100].
Figure 6. X-ray angiogram of the rabbit hindlimb prior to (A) superficial femoral artery occlusion and (B) post-occlusion via endovascular placement of platinum coils (black arrow). X-ray-visible microencapsulated stem cell injections injected intramuscularly in the medial thigh appear as radiopacities (white arrows). A quarter (Q) is used for reference measurements. Reprinted with permission from [151].
While these microencapsulation techniques, like direct labeling schemes, fail to measure cell viability, reporter gene labeling of the cells could be used to overcome this obstacle. At present, these microcapsules remain relatively large (~300–500 μm), therefore eliminating the possibility of direct intramyocardial or intracoronary injection. Furthermore, since the stem cells are trapped within the microcapsules, direct integration of the stem cells is prevented. Therefore, this technique will be most useful if stem cells are used to release cytokines to enhance angiogenesis and recruit native stem cells to differentiate into myocytes.
Conclusion
Stem cell labeling in conjunction with noninvasive imaging provides a powerful tool to aid in the optimization of stem cell type, selection dosing, delivery route and timing of transplantation to guide clinical cardiovascular stem cell trials. Despite significant progress in imaging techniques and label developments, no single labeling technique meets all the cardiac stem cell tracking criteria. A multimodality imaging approach is likely to play an important role in illuminating different aspects of stem cell biology in vivo and elucidating the mechanisms of cardiac repair and regeneration.

Future perspective
For now, stem cell labeling for noninvasive tracking will remain mostly a preclinical tool to obtain FDA approval for new stem cell biologics and to guide the design of clinical trials. Since most interventional procedures are performed with x-ray angiographic systems, fusion of x-ray imaging with CT, MRI, PET or echocardiography appears the mostly likely imaging platform for cardiac stem cell tracking in clinical settings in the next 3–5 years. Ultimately, ultrasound, optical or MRI tracking of stem cells will be adopted for serial tracking of stem cells owing to the lack of ionizing radiation. Thus, long-term, future efforts will focus on delivery using existing technologies with the development of multimodality imaging approaches to interpret the results from cardiac stem cell trials and assess the long-term effects of stem cell labeling.
Financial & competing interests disclosure
Grant support was received from the National Institutes of Health Heart, Lung and Blood Institute (NIHNHLBI- R21/R33 HL89029) and Maryland Stem Cell Research Foundation (2008-MD-SCRFII-0399). Dara L Kraitchman is a coinventor on a pending US patent pertaining to imaging-visible microcapsules and receives grant support from Siemens Healthcare. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
- Lloyd-Jones D, Adams RJ, Brown TM et al. Executive summary: heart disease and stroke statistics – 2010 update: a report from the American Heart Association. Circulation 121(7), 948–954 (2010).
- Rickers C, Gallegos R, Seethamraju RT et al. Applications of magnetic resonance imaging for cardiac stem cell therapy. J. Interv. Cardiol. 17(1), 37–46 (2004).
- Meyer GP, Wollert KC, Lotz J et al. Intracoronary bone marrow cell transfer after myocardial infarction. Eighteen months’ follow-up data from the randomized, controlled BOOST (bone marrow transfer to enhance ST-elevation infarct regeneration) trial. Circulation 113(10), 1287–1294 (2006).
- Meyer GP, Wollert KC, Lotz J et al. Intracoronary bone marrow cell transfer after myocardial infarction. 5-year follow-up from the randomized-controlled BOOST trial. Eur. Heart J. 30(24), 2978–2984 (2009).
- Cleland JG, Coletta AP, Abdellah AT et al. Clinical trials update from the American Heart Association 2006. OAT, SALT 1 and 2, MAGIC, ABCD, PABA-CHF, IMPROVECHF and percutaneous mitral annuloplasty. Eur. J. Heart Fail. 9(1), 92–97 (2007).
- Janssens S, Dubois C, Bogaert J et al. Autologous bone marrow-derived stem-cell transfer in patients with ST-segment elevation myocardial infarction: double-blind, randomised controlled trial. Lancet 367(9505), 113–121 (2006).
- Lunde K, Solheim S, Aakhus S, Arnesen H, Abdelnoor M, Forfang K. Autologous stem cell transplantation in acute myocardial infarction: the ASTAMI randomized controlled trial. Intracoronary transplantation of autologous mononuclear bone marrow cells, study design and safety aspects. Scand. Cardiovasc. J. 39(3), 150–158 (2005).
- Abdel-Latif A, Bolli R, Tleyjeh IM et al. Adult bone marrow-derived cells for cardiac repair: a systematic review and meta-analysis. Arch. Intern. Med. 167(10), 989–997 (2007).
- Fan L, Chen L, Chen X, Fu F. A meta-analysis of stem cell mobilization by granulocyte colony-stimulating factor in the treatment of acute myocardial infarction. Cardiovasc. Drugs Ther. 22(1), 45–54 (2008).
- Lipinski MJ, Biondi-Zoccai GG, Abbate A et al. Impact of intracoronary cell therapy on left ventricular function in the setting of acute myocardial infarction: a collaborative systematic review and meta-analysis of controlled clinical trials. J. Am. Coll. Cardiol. 50(18), 1761–1767 (2007).
- Singh S, Arora R, Handa K et al. Stem cells improve left ventricular function in acute myocardial infarction. Clin. Cardiol. 32(4), 176–180 (2009).
- Zhang SN, Sun AJ, Ge JB et al. Intracoronary autologous bone marrow stem cells transfer for patients with acute myocardial infarction. A meta-analysis of randomised controlled trials. Int. J. Cardiol. 136(2), 178–185 (2008).
- Frangioni JV, Hajjar RJ. In vivo tracking of stem cells for clinical trials in cardiovascular disease. Circulation 110(21), 3378–3383 (2004).
- Fu Y, Kraitchman DL. Stem cell labeling for noninvasive delivery and tracking in cardiovascular regenerative therapy. Expert Rev. Cardiovasc. Ther. 8(8), 1149–1160 (2010).
- Amsalem Y, Mardor Y, Feinberg MS et al. Iron-oxide labeling and outcome of transplanted mesenchymal stem cells in the infarcted myocardium. Circulation 116(Suppl. 11), 138–145 (2007).
- Henning TD, Boddington S, Daldrup-Link HE. Labeling hESCs and hMSCs with iron oxide nanoparticles for non-invasive in vivo tracking with MR imaging. J. Vis. Exp. 13, 685 (2008).
- Hill JM, Dick AJ, Raman VK et al. Serial cardiac magnetic resonance imaging of injected mesenchymal stem cells. Circulation 108(8), 1009–1014 (2003).
- Brenner W, Aicher A, Eckey T et al. 111In-labeled CD34+ hematopoietic progenitor cells in a rat myocardial infarction model. J. Nucl. Med. 45(3), 512–518 (2004).
- Doyle B, Kemp BJ, Chareonthaitawee P et al. Dynamic tracking during intracoronary injection of 18F-FDG-labeled progenitor cell therapy for acute myocardial infarction. J. Nucl. Med. 48(10), 1708–1714 (2007).
- Hoshino K, Ly HQ, Frangioni JV, Hajjar RJ. In vivo tracking in cardiac stem cell-based therapy. Prog. Cardiovasc. Dis 49(6), 414–420 (2007).
- Rosen AB, Kelly DJ, Schuldt AJ et al. Finding fluorescent needles in the cardiac haystack. tracking human mesenchymal stem cells labeled with quantum dots for quantitative in vivo three-dimensional fluorescence analysis. Stem Cells 25(8), 2128–2138 (2007).
- Cao F, Lin S, Xie X et al. In vivo visualization of embryonic stem cell survival, proliferation and migration after cardiac delivery. Circulation 113(7), 1005–1014 (2006).
- Naumova AV, Reinecke H, Yarnykh V, Deem J, Yuan C, Charles EM. Ferritin overexpression for noninvasive magnetic resonance imaging-based tracking of stem cells transplanted into the heart. Mol. Imag. 9(4), 201–210 (2010).
- Zen K, Okigaki M, Hosokawa Y et al. Myocardium-targeted delivery of endothelial progenitor cells by ultrasound-mediated microbubble destruction improves cardiac function via an angiogenic response. J. Mol. Cell. Cardiol. 40(6), 799–809 (2006).
- Kuliszewski MA, Fujii H, Liao C et al. Molecular imaging of endothelial progenitor cell engraftment using contrast-enhanced ultrasound and targeted microbubbles. Cardiovasc. Res. 83(4), 653–662 (2009).
- Bara C, Ghodsizad A, Niehaus M et al. In vivo echocardiographic imaging of transplanted human adult stem cells in the myocardium labeled with clinically applicable CliniMACS nanoparticles. J. Am. Soc. Echocardiogr. 19(5), 563–568 (2006).
- Shapiro EM, Skrtic S, Sharer K, Hill JM, Dunbar CE, Koretsky AP. MRI detection of single particles for cellular imaging. Proc. Natl Acad. Sci. USA 101(30), 10901–10906 (2004).
- Frank JA, Miller BR, Arbab AS et al. Clinically applicable labeling of mammalian and stem cells by combining superparamagnetic iron oxides and transfection agents. Radiology 228, 480–487 (2003).
- Walczak P, Kedziorek DA, Gilad AA, Lin S, Bulte JW. Instant MR labeling of stem cells using magnetoelectroporation. Magn. Reson. Med. 54(4), 769–774 (2005).
- Gilad AA, Walczak P, Mcmahon MT et al. MR tracking of transplanted cells with ‘positive contrast’ using manganese oxide nanoparticles. Magn. Reson. Med. 60(1), 1–7 (2008).
- Kim HS, Cho HR, Choi SH, Woo JS, Moon WK. In vivo imaging of tumor transduced with bimodal lentiviral vector encoding human ferritin and green fluorescent protein on a 1.5T clinical magnetic resonance scanner. Cancer Res. 70(18), 7315–7324 (2010).
- Moore A, Josephson L, Bhorade RM, Basilion JP, Weissleder R. Human transferrin receptor gene as a marker gene for MR imaging. Radiology 221(1), 244–250 (2001).
- Gerber BL, Raman SV, Nayak K et al. Myocardial first-pass perfusion cardiovascular magnetic resonance. history, theory and current state of the art. J. Cardiovasc. Magn. Reson. 10(1), 18 (2008).
- Adler ED, Bystrup A, Briley-Saebo KC et al. In vivo detection of embryonic stem cell-derived cardiovascular progenitor cells using Cy3-labeled Gadofluorine M in murine myocardium. JACC Cardiovasc. Imag. 2(9), 1114–1122 (2009).
- Kraitchman DL, Bulte JW. Imaging of stem cells using MRI. Basic Res. Cardiol. 103(2), 105–113 (2008).
- Bulte JW, Kraitchman DL. Monitoring cell therapy using iron oxide MR contrast agents. Curr. Pharm. Biotechnol. 5(6), 567–584 (2004).
- Bulte JW. In vivo MRI cell tracking: clinical studies. AJR Am. J. Roentgenol. 193(2), 314–325 (2009).
- Karussis D, Karageorgiou C, Vaknin- Dembinsky A et al. Safety and immunological effects of mesenchymal stem cell transplantation in patients with multiple sclerosis and amyotrophic lateral sclerosis. Arch. Neurol. 67(10), 1187–1194 (2010).
- De Vries IJ, Lesterhuis WJ, Barentsz JO et al. Magnetic resonance tracking of dendritic cells in melanoma patients for monitoring of cellular therapy. Nat. Biotechnol. 23(11), 1407–1413 (2005).
- Verdijk P, Scheenen TW, Lesterhuis WJ et al. Sensitivity of magnetic resonance imaging of dendritic cells for in vivo tracking of cellular cancer vaccines. Int. J. Cancer 120(5), 978–984 (2007).
- Zhu J, Zhou L, Xingwu F. Tracking neural stem cells in patients with brain trauma. N. Engl. J. Med. 355(22), 2376–2378 (2006).
- Ebert SN, Taylor DG, Nguyen HL et al. Noninvasive tracking of cardiac embryonic stem cells in vivo using magnetic resonance imaging techniques. Stem Cells 25(11), 2936–2944 (2007).
- Kraitchman DL, Heldman AW, Atalar E et al. In vivo magnetic resonance imaging of mesenchymal stem cells in myocardial infarction. Circulation 107(18), 2290–2293 (2003).
- Zhou R, Idiyatullin D, Moeller S et al. SWIFT detection of SPIO-labeled stem cells grafted in the myocardium. Magn. Reson. Med. 63(5), 1154–1161 (2010).
- Amado LC, Saliaris AP, Schuleri KH et al. Cardiac repair with intramyocardial injection of allogeneic mesenchymal stem cells after myocardial infarction. Proc. Natl Acad. Sci. USA 102(32), 11474–11479 (2005).
- Dick AJ, Guttman MA, Raman VK et al. Magnetic resonance fluoroscopy allows targeted delivery of mesenchymal stem cells to infarct borders in swine. Circulation 108(23), 2899–2904 (2003).
- Saeed M, Saloner D, Weber O, Martin A, Henk C, Higgins C. MRI in guiding and assessing intramyocardial therapy. Eur. Radiol. 15(5), 851–863 (2005).
- Kraitchman DL, Gilson WD, Lorenz CH. Stem cell therapy: MRI guidance and monitoring. J. Magn. Reson. Imag. 27(2), 299–310 (2008).
- Chen IY, Greve JM, Gheysens O et al. Comparison of optical bioluminescence reporter gene and superparamagnetic iron oxide MR contrast agent as cell markers for noninvasive imaging of cardiac cell transplantation. Mol. Imag. Biol. 11(3), 178–187 (2009).
- Terrovitis J, Stuber M, Youssef A et al. Magnetic resonance imaging overestimates ferumoxide-labeled stem cell survival after transplantation in the heart. Circulation 117(12), 1555–1562 (2008).
- Eibofner F, Steidle G, Kehlbach R, Bantleon R, Schick F. Positive contrast imaging of iron oxide nanoparticles with susceptibilityweighted imaging. Magn. Reson. Med. 64(4), 1027–1038 (2010).
- Cukur T, Yamada M, Overall WR, Yang P, Nishimura DG. Positive contrast with alternating repetition time SSFP (PARTS): a fast imaging technique for SPIO-labeled cells. Magn. Reson. Med. 63(2), 427–437 (2010).
- Stuber M, Gilson WD, Schar M et al. Positive contrast visualization of iron oxide-labeled stem cells using inversion-recovery with ON-resonant water suppression (IRON). Magn. Reson. Med. 58(5), 1072–1077 (2007).
- Ahrens ET, Flores R, Xu H, Morel PA. In vivo imaging platform for tracking immunotherapeutic cells. Nat. Biotechnol. 23(8), 983–987 (2005).
- Barnett BP, Ruiz-Cabello J, Hota P et al. Fluorocapsules for improved function, immunoprotection and visualization of cellular therapeutics with MR, US and CT imaging. Radiology 258(1), 182–191 (2010).
- Fu Y, Kedziorek D, Ouwerkerk R et al. Multifunctional perfluorooctylbromide alginate microcapsules for monitoring of mesenchymal stem cell delivery using CT and MRI. J. Cardiovasc. Magn. Reson. 11(Suppl. 1), O7 (2009).
- Partlow KC, Chen J, Brant JA et al. 19F magnetic resonance imaging for stem/ progenitor cell tracking with multiple unique perfluorocarbon nanobeacons. FASEB J. 21(8), 1647–1654 (2007).
- Bulte JW. Hot spot MRI emerges from the background. Nat. Biotechnol. 23(8), 945–946 (2005)
- Koretsky AP, Brosnan MJ, Chen LH, Chen JD, Van Dyke T. NMR detection of creatine kinase expressed in liver of transgenic mice: determination of free ADP levels. Proc. Natl Acad. Sci. USA 87(8), 3112–3116 (1990).
- Liu J, Cheng EC, Long Jr RC et al. Noninvasive monitoring of embryonic stem cells in vivo with MRI transgene reporter. Tissue Eng. Part C Methods 15(4), 739–747 (2009).
- Deans AE, Wadghiri YZ, Bernas LM, Yu X, Rutt BK, Turnbull DH. Cellular MRI contrast via coexpression of transferrin receptor and ferritin. Magn. Reson. Med. 56(1), 51–59 (2006).
- Pawelczyk E, Arbab AS, Pandit S, Hu E, Frank JA. Expression of transferrin receptor and ferritin following ferumoxides–protamine sulfate labeling of cells: implications for cellular magnetic resonance imaging. NMR Biomed. 19(5), 581–592 (2006).
- Genove G, Demarco U, Xu H, Goins WF, Ahrens ET. A new transgene reporter for in vivo magnetic resonance imaging. Nat. Med. 11(4), 450–454 (2005).
- Gilad AA, McMahon MT, Walczak P et al. Artificial reporter gene providing MRI contrast based on proton exchange. Nat. Biotechnol. 25(2), 217–219 (2007).
- Massoud TF, Gambhir SS. Molecular imaging in living subjects: seeing fundamental biological processes in a new light. Genes Dev. 17(5), 545–580 (2003).
- Kendziorra K, Barthel H, Erbs S et al. Effect of progenitor cells on myocardial perfusion and metabolism in patients after recanalization of a chronically occluded coronary artery. J. Nucl. Med. 49(4), 557–563 (2008).
- Castellani M, Colombo A, Giordano R et al. The role of PET with 13N-ammonia and 18F-FDG in the assessment of myocardial perfusion and metabolism in patients with recent AMI and intracoronary stem cell injection. J. Nucl. Med. 51(12), 1908–1916 (2010).
- Kraitchman DL, Tatsumi M, Gilson WD et al. Dynamic imaging of allogeneic mesenchymal stem cells trafficking to myocardial infarction. Circulation 112(10), 1451–1461 (2005).
- Aicher A, Brenner W, Zuhayra M et al. Assessment of the tissue distribution of transplanted human endothelial progenitor cells by radioactive labeling. Circulation 107(16), 2134–2139 (2003).
- Zhou R, Thomas DH, Qiao H et al. In vivo detection of stem cells grafted in infarcted rat myocardium. J. Nucl. Med. 46(5), 816–822 (2005).
- Brenner W, Aicher A, Eckey T et al. 111In-labeled CD34+ hematopoietic progenitor cells in a rat myocardial infarction model. J. Nucl. Med. 45(3), 512–518 (2004).
- Hou D, Youssef EA, Brinton TJ et al. Radiolabeled cell distribution after intramyocardial, intracoronary and interstitial retrograde coronary venous delivery. implications for current clinical trials. Circulation 112(Suppl. 9) I150–I156 (2005).
- Tran N, Li Y, Maskali F et al. Short-term heart retention and distribution of intramyocardial delivered mesenchymal cells within necrotic or intact myocardium. Cell Transplant. 15(4), 351–358 (2006).
- Lyngbaek S, Ripa RS, Haack-Sorensen M et al. Serial in vivo imaging of the porcine heart after percutaneous, intramyocardially injected 111In-labeled human mesenchymal stromal cells. Int. J. Cardiovasc. Imag. 26(3), 273–284 (2010).
- Wisenberg G, Lekx K, Zabel P et al. Cell tracking and therapy evaluation of bone marrow monocytes and stromal cells using SPECT and CMR in a canine model of myocardial infarction. J. Cardiovasc. Magn. Reson. 11(1), 11 (2009).
- Blackwood KJ, Lewden B, Wells RG et al. In vivo SPECT quantification of transplanted cell survival after engraftment using 111In-tropolone in infarcted canine myocardium. J. Nucl. Med. 50(6), 927–935 (2009).
- Caveliers V, De Keulenaer G, Everaert H et al. In vivo visualization of 111In labeled CD133+ peripheral blood stem cells after intracoronary administration in patients with chronic ischemic heart disease. Q. J. Nucl. Med. Mol. Imag. 51(1), 61–66 (2007).
- Schots R, De Keulenaer G, Schoors D et al. Evidence that intracoronary-injected CD133+ peripheral blood progenitor cells home to the myocardium in chronic postinfarction heart failure. Exp. Hematol. 35(12), 1884–1890 (2007).
- Hofmann M, Wollert KC, Meyer GP et al. Monitoring of bone marrow cell homing into the infarcted human myocardium. Circulation 111(17), 2198–2202 (2005).
- Kang WJ, Kang HJ, Kim HS, Chung JK, Lee MC, Lee DS. Tissue distribution of 18F-FDG-labeled peripheral hematopoietic stem cells after intracoronary administration in patients with myocardial infarction. J. Nucl. Med. 47(8), 1295–1301 (2006).
- Tran N, Franken PR, Maskali F et al. Intramyocardial implantation of bone marrow-derived stem cells enhances perfusion in chronic myocardial infarction. Dependency on initial perfusion depth and follow-up assessed by gated pinhole SPECT. J. Nucl. Med. 48(3), 405–412 (2007).
- Jin Y, Kong H, Stodilka RZ et al. Determining the minimum number of detectable cardiac-transplanted 111Intropolone- labelled bone-marrow-derived mesenchymal stem cells by SPECT. Phys. Med. Biol. 50(19), 4445–4455 (2005).
- Gildehaus FJ, Haasters F, Drosse I et al. Impact of indium-111 oxine labelling on viability of human mesenchymal stem cells in vitro and 3D cell-tracking using SPECT/ CT in vivo. Mol. Imag. Biol. DOI: 10.1007/ s11307–010–0439–1 (2010) (Epub ahead of print).
- Gholamrezanezhad A, Mirpour S, Ardekani JM et al. Cytotoxicity of 111In-oxine on mesenchymal stem cells. A time-dependent adverse effect. Nucl. Med. Commun. 30(3), 210–216 (2009).
- Aicher A, Brenner W, Zuhayra M et al. Assessment of the tissue distribution of transplanted human endothelial progenitor cells by radioactive labeling. Circulation 107(16), 2134–2139 (2003).
- Lee Z, Dennis JE, Gerson SL. Imaging stem cell implant for cellular-based therapies. Exp. Biol. Med.(Maywood) 233(8), 930–940 (2008).
- Miyagawa M, Beyer M, Wagner B et al. Cardiac reporter gene imaging using the human sodium/iodide symporter gene. Cardiovasc. Res. 65(1), 195–202 (2005).
- Qiao H, Zhang H, Yamanaka S et al. Long-term improvement in postinfarct left ventricular global and regional contractile function is mediated by embryonic stem cell-derived cardiomyocytes. Circ. Cardiovasc. Imag. 4(1), 33–41 (2011).
- Sun X, Annala AJ, Yaghoubi SS et al. Quantitative imaging of gene induction in living animals. Gene Ther. 8(20), 1572–1579 (2001).
- Yaghoubi SS, Wu L, Liang Q et al. Direct correlation between positron emission tomographic images of two reporter genes delivered by two distinct adenoviral vectors. Gene Ther. 8(14), 1072–1080 (2001).
- Gyongyosi M, Blanco J, Marian T et al. Serial noninvasive in vivo positron emission tomographic tracking of percutaneously intramyocardially injected autologous porcine mesenchymal stem cells modified for transgene reporter gene expression. Circ. Cardiovasc. Imag. 1(2), 94–103 (2008).
- Luker GD, Sharma V, Pica CM et al. Noninvasive imaging of protein–protein interactions in living animals. Proc. Natl Acad. Sci. USA 99(10), 6961–6966 (2002).
- Zhang SJ, Wu JC. Comparison of imaging techniques for tracking cardiac stem cell therapy. J. Nucl. Med. 48(12), 1916–1919 (2007).
- Gruber PJ, Li Z, Li H et al. In vivo imaging of MLC2v-luciferase, a cardiac-specific reporter gene expression in mice. Acad. Radiol. 11(9), 1022–1028 (2004).
- Fu Y, Kraitchman DL. Stem cell labeling for noninvasive delivery and tracking in cardiovascular regenerative therapy. Expert Rev. Cardiovasc. Ther. 8(8), 1149–1160 (2010).
- Ransohoff KJ, Wu JC. Advances in cardiovascular molecular imaging for tracking stem cell therapy. Thromb. Haemost. 104(1), 13–22 (2010).
- De Silva R, Gutierrez LF, Raval AN, McVeigh ER, Ozturk C, Lederman RJ. X-ray fused with magnetic resonance imaging (XFM) to target endomyocardial injections. Validation in a swine model of myocardial infarction. Circulation 114(22), 2342–2350 (2006).
- Barnett BP, Arepally A, Karmarkar PV et al. Magnetic resonance-guided, real-time targeted delivery and imaging of magnetocapsules immunoprotecting pancreatic islet cells. Nat. Med. 13(8), 986–991 (2007).
- Barnett BP, Kraitchman DL, Lauzon C et al. Radiopaque alginate microcapsules for x-ray visualization and immunoprotection of cellular therapeutics. Mol. Pharm. 3(5), 531–538 (2006).
- Gilson WD, Kraitchman DL. Noninvasive cardiovascular imaging techniques for basic science research: application to cellular therapeutics. Rev. Esp. Cardiol. 62(8), 918–927 (2009).
- Nahrendorf M, Sosnovik D, French B et al. Multimodality cardiovascular molecular imaging - Part II. Circ. Cardiovasc. Imag. 2, 56–70 (2009).
- Drexler H, Meyer GP, Wollert KC. Bone-marrow-derived cell transfer after ST-elevation myocardial infarction: lessons from the BOOST trial. Nat. Clin. Pract. Cardiovasc. Med. 3(Suppl. 1), S65–S68 (2006).
- Schaefer A, Meyer GP, Fuchs M et al. Impact of intracoronary bone marrow cell transfer on diastolic function in patients after acute myocardial infarction: results from the BOOST trial. Eur. Heart J. 27(8), 929–935 (2006).
- Schaefer A, Zwadlo C, Fuchs M et al. Long-term effects of intracoronary bone marrow cell transfer on diastolic function in patients after acute myocardial infarction. 5-year results from the randomized-controlled BOOST trial – an echocardiographic study. Eur. J. Echocardiogr. 11(2), 165–171 (2010).
- Wollert KC, Meyer GP, Lotz J et al. Intracoronary autologous bone-marrow cell transfer after myocardial infarction. The BOOST randomised controlled clinical trial. Lancet 364(9429), 141–148 (2004).
- Mansour S, Roy DC, Bouchard V et al. COMPARE-AMI trial. Comparison of intracoronary injection of CD133+ bone marrow stem cells to placebo in patients after acute myocardial infarction and left ventricular dysfunction: study rationale and design. J. Cardiovasc. Transl. Res. 3(2), 153–159 (2010).
- Schachinger V, Erbs S, Elsasser A et al. Intracoronary bone marrow-derived progenitor cells in acute myocardial infarction. N. Engl. J. Med. 355(12), 1210–1221 (2006).
- Tendera M, Wojakowski W, Ruzyllo W et al. Intracoronary infusion of bone marrowderived selected CD34+CXCR4+ cells and non-selected mononuclear cells in patients with acute STEMI and reduced left ventricular ejection fraction. Results of randomized, multicentre Myocardial Regeneration by Intracoronary Infusion of Selected Population of Stem Cells in Acute Myocardial Infarction (REGENT) Trial. Eur. Heart J. 30(11), 1313–1321 (2009).
- Tse HF, Thambar S, Kwong YL et al. Prospective randomized trial of direct endomyocardial implantation of bone marrow cells for treatment of severe coronary artery diseases (PROTECT-CAD trial). Eur. Heart J. 28(24), 2998–3005 (2007).
- Chan CW, Kwong YL, Kwong RY, Lau CP, Tse HF. Improvement of myocardial perfusion reserve detected by cardiovascular magnetic resonance after direct endomyocardial implantation of autologous bone marrow cells in patients with severe coronary artery disease. J. Cardiovasc. Magn. Reson. 12, 6 (2010).
- Brehm M, Strauer BE. Stem cell therapy in postinfarction chronic coronary heart disease. Nat. Clin. Pract. Cardiovasc. Med. 3(Suppl. 1), S101–S104 (2006).
- Fernandez-Aviles F, San Roman JA, Garcia-Frade J et al. Experimental and clinical regenerative capability of human bone marrow cells after myocardial infarction. Circ. Res. 95(7), 742–748 (2004).
- Fuchs S, Satler LF, Kornowski R et al. Catheter-based autologous bone marrow myocardial injection in no-option patients with advanced coronary artery disease. a feasibility study. J. Am. Coll. Cardiol. 41(10), 1721–1724 (2003).
- Galinanes M, Loubani M, Davies J, Chin D, Pasi J, Bell PR. Autotransplantation of unmanipulated bone marrow into scarred myocardium is safe and enhances cardiac function in humans. Cell Transplant. 13(1), 7–13 (2004).
- Hamano K, Nishida M, Hirata K et al. Local implantation of autologous bone marrow cells for therapeutic angiogenesis in patients with ischemic heart disease: clinical trial and preliminary results. Jpn Circ. J. 65(9), 845–847 (2001).
- Strauer BE, Yousef M, Schannwell CM. The acute and long-term effects of intracoronary stem cell transplantation in 191 patients with chronic heart failure: the STAR-heart study. Eur. J. Heart Fail. 12(7), 721–729 (2010).
- Strauer BE, Brehm M, Zeus T et al. Regeneration of human infarcted heart muscle by intracoronary autologous bone marrow cell transplantation in chronic coronary artery disease. The IACT Study. J. Am. Coll. Cardiol. 46(9), 1651–1658 (2005).
- Kuethe F, Richartz BM, Sayer HG et al. Lack of regeneration of myocardium by autologous intracoronary mononuclear bone marrow cell transplantation in humans with large anterior myocardial infarctions. Int. J. Cardiol. 97(1), 123–127 (2004).
- Lunde K, Solheim S, Aakhus S et al. Intracoronary injection of mononuclear bone marrow cells in acute myocardial infarction. N. Engl. J. Med. 355(12), 1199–1209 (2006).
- Perin EC, Dohmann HF, Borojevic R et al. Transendocardial, autologous bone marrow cell transplantation for severe, chronic ischemic heart failure. Circulation 107(18), 2294–2302 (2003).
- Perin EC, Dohmann HF, Borojevic R et al. Improved exercise capacity and ischemia 6 and 12 months after transendocardial injection of autologous bone marrow mononuclear cells for ischemic cardiomyopathy. Circulation 110(11 Suppl. 1), II213–II218 (2004).
- Silva GV, Perin EC, Dohmann HF et al. Catheter-based transendocardial delivery of autologous bone-marrow-derived mononuclear cells in patients listed for heart transplantation. Tex. Heart. Inst. J. 31(3), 214–219 (2004).
- Arnold R, Villa A, Gutierrez H et al. Absence of accelerated atherosclerotic disease progression after intracoronary infusion of bone marrow derived mononuclear cells in patients with acute myocardial infarction – angiographic and intravascular ultrasound – results from the TErapia Celular Aplicada al Miocardio Pilot study. Am. Heart J. 159(6), 1154, E1151–E1158 (2010).
- van Ramshorst J, Bax JJ, Beeres SL et al. Intramyocardial bone marrow cell injection for chronic myocardial ischemia: a randomized controlled trial. JAMA 301(19), 1997–2004 (2009).
- Strauer BE, Brehm M, Zeus T et al. Repair of infarcted myocardium by autologous intracoronary mononuclear bone marrow cell transplantation in humans. Circulation 106(15), 1913–1918 (2002).
- Willerson JT, Perin EC, Ellis SG et al. Intramyocardial injection of autologous bone marrow mononuclear cells for patients with chronic ischemic heart disease and left ventricular dysfunction (First Mononuclear Cells injected in the US [FOCUS]). Rationale and design. Am. Heart J. 160(2), 215–223 (2010).
- Assmus B, Honold J, Schachinger V et al. Transcoronary transplantation of progenitor cells after myocardial infarction. N. Engl. J. Med. 355(12), 1222–1232 (2006).
- Assmus B, Schachinger V, Teupe C et al. Transplantation of Progenitor Cells and Regeneration Enhancement in Acute Myocardial Infarction (TOPCARE-AMI). Circulation 106(24), 3009–3017 (2002).
- Britten MB, Abolmaali ND, Assmus B et al. Infarct remodeling after intracoronary progenitor cell treatment in patients with acute myocardial infarction (TOPCAREAMI). Mechanistic insights from serial contrast-enhanced magnetic resonance imaging. Circulation 108(18), 2212–2218 (2003).
- Schachinger V, Assmus B, Britten MB et al. Transplantation of progenitor cells and regeneration enhancement in acute myocardial infarction. Final one-year results of the TOPCARE-AMI trial. J. Am. Coll. Cardiol. 44(8), 1690–1699 (2004).
- Bartunek J, Vanderheyden M, Vandekerckhove B et al. Intracoronary injection of CD133-positive enriched bone marrow progenitor cells promotes cardiac recovery after recent myocardial infarction. feasibility and safety. Circulation 112(9 Suppl.), I178–I183 (2005).
- Goussetis E, Manginas A, Koutelou M et al. Intracoronary infusion of CD133+ and CD133-CD34+ selected autologous bone marrow progenitor cells in patients with chronic ischemic cardiomyopathy. Cell isolation, adherence to the infarcted area and body distribution. Stem Cells 24(10), 2279–2283 (2006).
- Stamm C, Kleine HD, Westphal B et al. CABG and bone marrow stem cell transplantation after myocardial infarction. Thorac. Cardiovasc. Surg. 52(3), 152–158 (2004).
- Stamm C, Westphal B, Kleine HD et al. Autologous bone-marrow stem-cell transplantation for myocardial regeneration. Lancet 361(9351), 45–46 (2003).
- Losordo DW, Schatz RA, White CJ et al. Intramyocardial transplantation of autologous CD34+ stem cells for intractable angina. A Phase I/IIa double-blind, randomized controlled trial. Circulation 115(25), 3165–3172 (2007).
- Choi JH, Choi J, Lee WS et al. Lack of additional benefit of intracoronary transplantation of autologous peripheral blood stem cell in patients with acute myocardial infarction. Circ. J. 71(4), 486–494 (2007).
- Kang HJ, Lee HY, Na SH et al. Differential effect of intracoronary infusion of mobilized peripheral blood stem cells by granulocyte colony-stimulating factor on left ventricular function and remodeling in patients with acute myocardial infarction versus old myocardial infarction. The MAGIC Cell-3-DES randomized, controlled trial. Circulation 114(Suppl. 1), I145–I151 (2006).
- Chachques JC, Herreros J, Trainini J et al. Autologous human serum for cell culture avoids the implantation of cardioverterdefibrillators in cellular cardiomyoplasty. Int. J. Cardiol. 95(Suppl. 1), S29–S33 (2004).
- Dib N, Michler RE, Pagani FD et al. Safety and feasibility of autologous myoblast transplantation in patients with ischemic cardiomyopathy. Four-year follow-up. Circulation 112(12), 1748–1755 (2005).
- Herreros J, Prosper F, Perez A et al. Autologous intramyocardial injection of cultured skeletal muscle-derived stem cells in patients with non-acute myocardial infarction. Eur. Heart J. 24(22), 2012–2020 (2003).
- Ince H, Petzsch M, Rehders TC, Chatterjee T, Nienaber CA. Transcatheter transplantation of autologous skeletal myoblasts in postinfarction patients with severe left ventricular dysfunction. J. Endovasc. Ther. 11(6), 695–704 (2004).
- Menasche P, Hagege AA, Vilquin JT et al. Autologous skeletal myoblast transplantation for severe postinfarction left ventricular dysfunction. J. Am. Coll. Cardiol. 41(7), 1078–1083 (2003).
- Siminiak T, Fiszer D, Jerzykowska O et al. Percutaneous trans-coronary-venous transplantation of autologous skeletal myoblasts in the treatment of post-infarction myocardial contractility impairment. the POZNAN trial. Eur. Heart J. 26(12), 1188–1195 (2005).
- Siminiak T, Kalawski R, Fiszer D et al. Autologous skeletal myoblast transplantation for the treatment of postinfarction myocardial injury. Phase I clinical study with 12 months of follow-up. Am. Heart J. 148(3), 531–537 (2004).
- Smits PC, Van Geuns RJ, Poldermans D et al. Catheter-based intramyocardial injection of autologous skeletal myoblasts as a primary treatment of ischemic heart failure. Clinical experience with six-month follow-up. J. Am. Coll. Cardiol. 42(12), 2063–2069 (2003).
- Chen SL, Fang WW, Ye F et al. Effect on left ventricular function of intracoronary transplantation of autologous bone marrow mesenchymal stem cell in patients with acute myocardial infarction. Am. J. Cardiol. 94(1), 92–95 (2004).
- Chen S, Liu Z, Tian N et al. Intracoronary transplantation of autologous bone marrow mesenchymal stem cells for ischemic cardiomyopathy due to isolated chronic occluded left anterior descending artery. J. Invasive Cardiol. 18(11), 552–556 (2006).
- Hare JM, Traverse JH, Henry TD et al. A randomized, double-blind, placebocontrolled, dose-escalation study of intravenous adult human mesenchymal stem cells (prochymal) after acute myocardial infarction. J. Am. Coll. Cardiol. 54(24), 2277–2286 (2009).
- Katritsis DG, Sotiropoulou PA, Karvouni E et al. Transcoronary transplantation of autologous mesenchymal stem cells and endothelial progenitors into infarcted human myocardium. Catheter Cardiovasc. Interv. 65(3), 321–329 (2005).
- Lasala GP, Silva JA, Kusnick BA, minguell JJ. Combination stem cell therapy for the treatment of medically refractory coronary ischemia: a Phase I study. Cardiovasc. Revasc. Med. 12(1), 29–34 (2011).
- Nahrendorf M, Sosnovik DE, French BA et al. Multimodality cardiovascular molecular imaging, Part II. Circ. Cardiovasc. Imag. 2(1), 56–70 (2009).